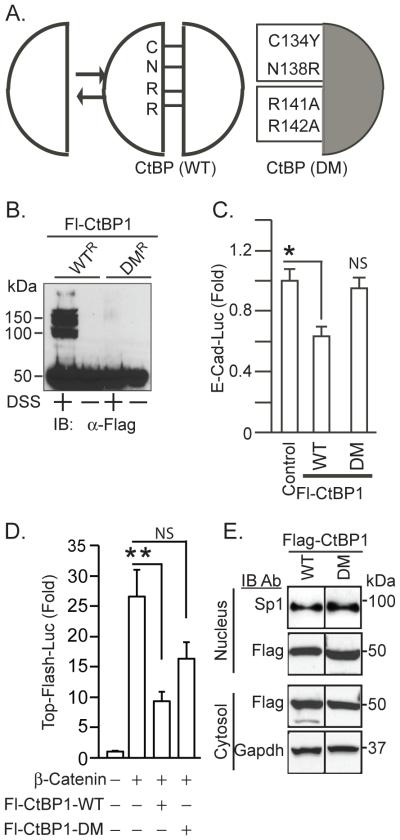
Fig. 1.
Mutations in the CtBP dimerization interface prevent oligomerization. (A) amino acid substitutions in the CtBP dimerization mutants. (B) Inability of the mutants to oligomerize. HuTu 80 cells depleted of endogenous CtBP stably expressing Fl-CtBP1-WTR or Fl-CtBP1-DMR were cross-linked with (+) or without (−) 5 mM disuccinimidyl suberate (DSS) for 30 mins. The expressed proteins were detected by immunoblotting with anti-Flag. (C) Monomeric CtBP does not repress E-Cadherin gene transcription. HuTu 80 cells were co-transfected with an E-Cadherin-Luciferase reporter, plus expression plasmids for wild type or mutant Fl-CtBP1. Luciferase activity normalized for transfection efficiency was measured 24 h after transfection. (D) Oligomeric but not monomeric CtBP represses ß catenin dependent transcription. HEK293 cells were co-transfected with TOPflash reporter in the absence (−) or the presence (+) of the indicated expression plasmids. (C, D) Results shown as the mean ± SEM (n=5) *P<0.02, ** P <0.01, NS, not significant. (E) Distribution of wild type and dimerization mutants of Fl-CtBP1, between the cytosol and nucleus. Cytoplasmic and nuclear extracts were prepared from HuTu 80 cells transfected pcDNA-Fl-CtBP1 (WT or DM) and were analyzed for expression of Flag proteins by immunoblotting. Immunoblotting for GAPDH and Sp1 served as loading controls for cytoplasmic and nuclear proteins respectively.