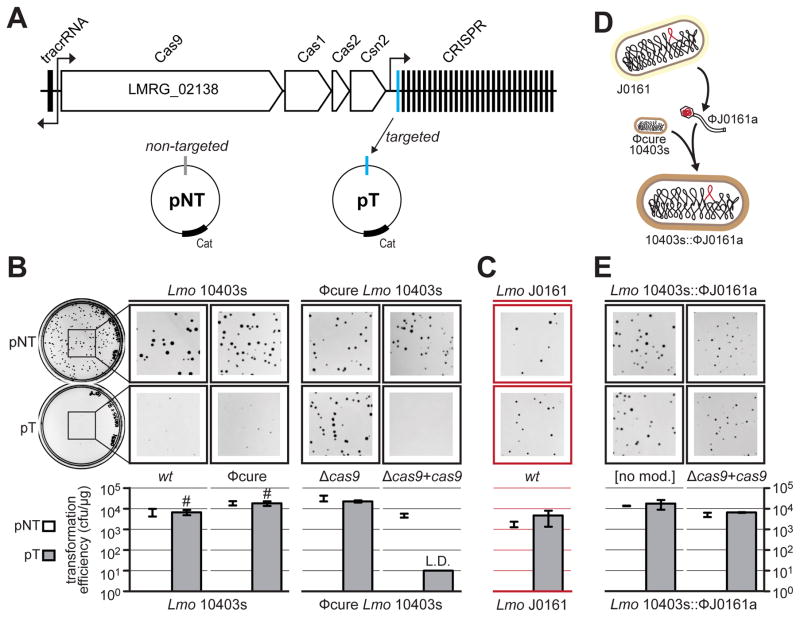
Figure 2. A prophage from L. monocytogenes J0161 inhibits CRISPR-Cas9 function.
(A) The type II-A CRISPR-Cas locus in L. monocytogenes 10403s. Four cas genes and the upstream tracrRNA are indicated, along with a CRISPR array containing 30 spacers. The predicted direction of transcription is indicated with black arrows. Subsequent experiments utilize a non-targeted plasmid (pNT) and a targeted plasmid (pT) that has a protospacer matching spacer 1 in this strain.
(B) Representative pictures of colonies of Lmo 10403s wild type (wt), prophage-cured (ϕcure), cas9-deletion strain (Δcas9), and a cas9 overexpression strain (Δcas9 + cas9) after being transformed with pT or pNT plasmids. Bar graphs below the plates show the calculated transformation efficiency (colony forming units per μg of plasmid). Data are represented as the mean of three biological replicates +/− SD. L.D. limit of detection, transformants with small colonies denoted with #.
(C) Plasmid-targeting assay with wild type J0161 (contains the ϕJ0161a prophage; experiment conducted as in (B), except with pTJ0161 as the targeted plasmid) is shown in red to denote self-targeting (as in Figure 1).
(D) A schematic demonstrating the construction of a 10403s strain containing the prophage ϕJ0161a (10403s::ϕJ0161a). See STAR Methods for details. (E) Plasmid-targeting assay with 10403s lysogenized with the ϕJ0161a prophage (10403s::ϕJ0161a) with endogenous (no mod) or overexpressed cas9 (Δcas9 + cas9; experiment conducted as in (B)).