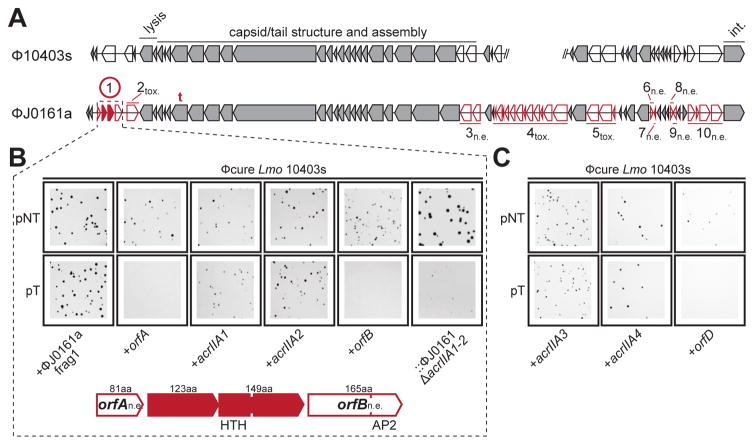
Figure 3. Identification of four distinct anti-CRISPR proteins.
(A) Comparison of the open reading frames from two similar prophages from L. monocytogenes 10403s and J0161. Unique genes (red) comprising ten fragments of ϕJ0161 were tested for CRISPR-Cas9 inhibition in 10403s. n.e., No effect on CRISPR-Cas9 activity, tox., fragment toxic when expressed, t., location of self-targeted protospacer. The encircled fragment exhibited anti-CRISPR activity with two genes (acrAII1, acrAII2) independently capable of inhibiting CRISPR-Cas activity. Conserved (grey) genes were not tested. For reference, phage genes involved in cell lysis, capsid assembly and host integration (int.) are labeled.
(B) Representative colony pictures of Lmo 10403s ϕcure strains constitutively expressing “fragment 1” (as shown in (A)) or the indicated individual genes from ϕJ0161a transformed with pNT or pT. The rightmost panels show a 10403s lysogen of ϕJ0161a with CRISPR-Cas9 inhibitor genes deleted (::ϕJ0161aΔacrIIA1-2). See Figure S2 for data from the other ϕJ0161a fragments and Figure S2/S3 for full plates.
(C) Representative colony pictures of Lmo 10403s ϕcure strains constitutively expressing acrIIA3, acrIIA4, or orfD transformed with pNT or pT.