Figure 6. The LIR Domain of PHB2 Is Required for Parkin-Mediated Mitophagy.
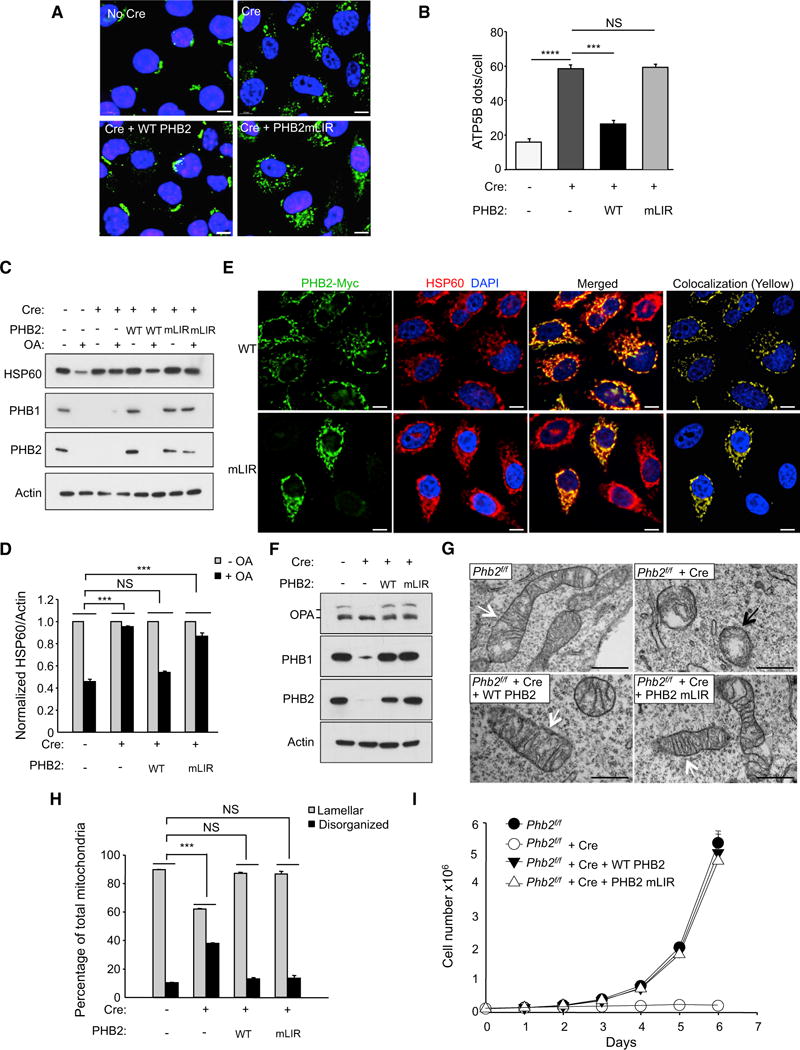
(A) Representative images of ATP5B immunofluorescence staining of Phb2flox/flox/Parkin MEFs transduced with empty vector or a Cre-expressing lentivirus and treated with OA for 16 hr prior to imaging and automated image analysis. Scale bars, 10 μm.
(B) Quantitation of cytoplasmic ATP5B puncta in Phb2flox/flox/Parkin MEFs transduced with lentiviruses expressing proteins indicated below x axis. Shown are mean ± SEM of 150 cells analyzed per condition. Similar results were observed in three independent experiments. ***p < 0.001; NS, not significant; one-way ANOVA with the Sidak test.
(C) Western blot analysis of indicated mitophagy markers in MEFs treated as shown in (A).
(D) Densitometric quantification of HSP60/Actin in (C). Shown are mean ± SEM values from western blot analyses of three independent experiments. NS, not significant; ***p < 0.001; two-way ANOVA for comparison of magnitude of change ± OA in indicated group versus Cre (−) control group.
(E) Representative images showing co-localization of PHB2-myc and HSP60 in HeLa/Parkin cells. Twenty-four hours prior to experiment, endogenous PHB2 and PHB1 were depleted with PHB2 siRNA and cells were co-transfected with indicated PHB2-myc construct and non-tagged PHB1. Scale bars, 10 μm.
(F) Western blot analysis to detect OPA1 processing Phb2flox/flox/Parkin MEFs during normal growth conditions with or without expression of Cre and WT PHB2 or PHB2 mLIR. Similar results were observed in two independent experiments.
(G) Representative images of mitochondria in MEFs shown in (F) that would be classified as lamellar (white arrows) or disorganized (black arrows).
(H) Quantification of mitochondrial cristae morphology for MEFs shown in (F) and (G). For each genotype, mitochondria were scored in 50 cell profiles. Bars are mean ± SEM for triplicate samples per genotype. NS, not significant, ***p < 0.001; two-way ANOVA for comparison of magnitude of difference between lamellar versus disorganized mitochondria in indicated group as compared to Cre (−) control group.
(I) Growth curves of MEFS of indicated genotype. Cells (2 × 104) were plated and counted at serial time points. Bars are mean ± SEM of triplicate samples for each genotype at each time point. p < 0.0001 for Phb2f/f + Cre versus all other groups; liner mixed-model effect. No differences were observed between Phb2f/f + Cre + WT PHB2 versus Phb2f/f + Cre + PHB2 mLIR.
