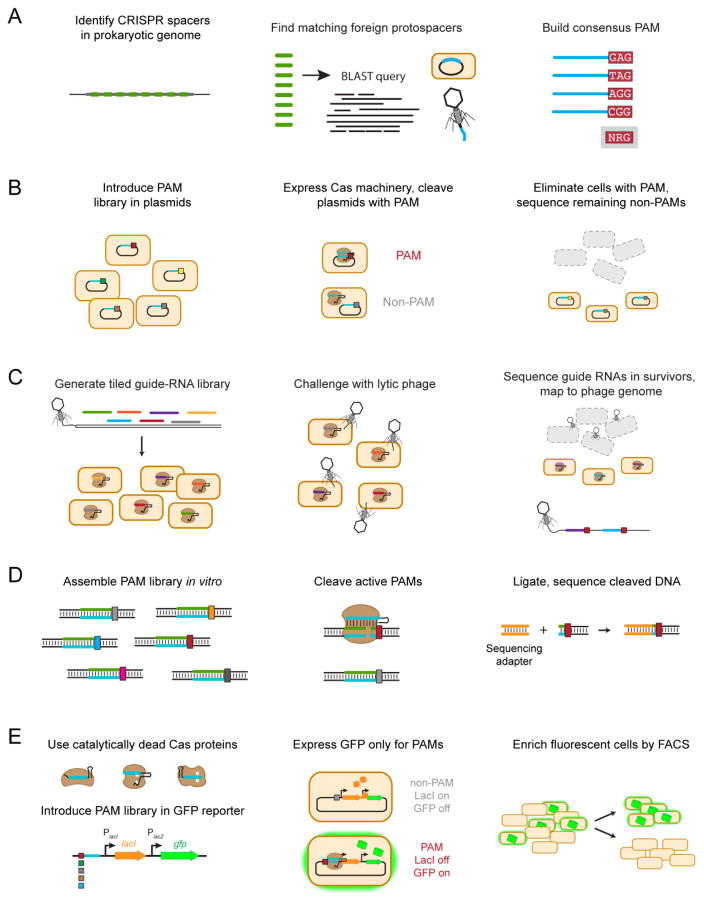
Figure 4.
Methods for PAM determination. (A) In silico PAM determination. A BLAST search of metagenomic databases identifies potential protospacers. Matches found on foreign nucleic acid elements from bacteriophages and plasmids are aligned to elucidate a single consensus sequence. (B) PAM determination through plasmid clearance. Plasmids harboring a library of potential PAM sequences are transformed into cells expressing the Cas proteins and the targeting guide RNA, and functional PAM sequences are identified based on their depletion from the library. (C) PAM determination through bacteriophage clearance. A library of guide RNAs are designed so their targets tile along a lytic bacteriophage genome. Guide RNAs targeting a site flanked by a functional PAM protect the bacteriophage attack, as revealed by sequencing the enriched guide RNAs and mapping their locations and flanking sequences within the bacteriophage genome. (D) In vitro PAM determination through DNA cleavage. A large PAM library is incubated with purified Cas proteins and transcribed crRNAs in a reaction buffer. Functional PAMs within this library are cleaved, and a sequencing adapter (shown in orange) can then be ligated for high-throughput sequencing. Alternatively, intact DNA can be sequenced, revealing PAM sequences that were depleted because of DNA cleavage. (E) In vivo PAM determination through DNA binding. Catalytically-dead effector proteins are targeted to a PAM library upstream of the lac operon regulating GFP expression. If a functional PAM sequence is present, binding by the catalytically-dead effector proteins represses expression of LacI, resulting in de-repression of GFP. Fluorescence cells are then isolated by fluorescent-activated cell sorting and sequenced.