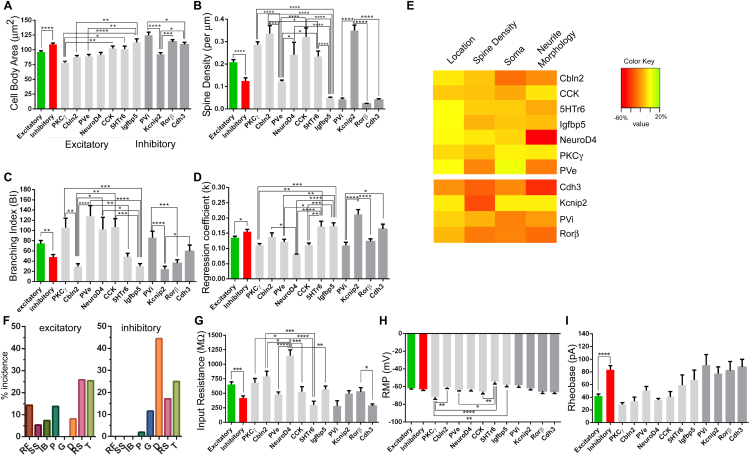
Figure S4.
Additional Morphometric and Physiological Characterization of 11 Interneurons of the LTMR-RZ, Related to Figures 3 and 4
(A) Cell body area summary for excitatory and inhibitory subtypes. For excitatory versus inhibitory comparison: (unpaired t test ∗∗∗∗p < 0.0001). For excitatory group: (one-way ANOVA: p < 0.0001, F[6,201] = 6.562). For inhibitory group: (one-way ANOVA: p < 0.0001, F[3,142] = 12.47). Post hoc Tukey’s test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
(B) Spine density measurements for excitatory and inhibitory subtypes. For excitatory versus inhibitory comparison: (unpaired t test ∗∗p < 0.0001). For excitatory group: (one-way ANOVA: p < 0.0001, F[6,187] = 24.39). For inhibitory group: (one-way ANOVA: p < 0.0001, F[3,125] = 132.1). Post hoc Tukey’s test: ∗p < 0.05, ∗∗∗∗p < 0.0001.
(C) Branching index (BI) summary describing ramification patterns for excitatory and inhibitory subtypes. BI values are positively correlated to branching complexity. For excitatory versus inhibitory comparison: (unpaired t test ∗∗p < 0.005). For excitatory group: (one-way ANOVA: p < 0.0001, F[6,194] = 9.207). For inhibitory group: (one-way ANOVA: p < 0.0001, F[3,138] = 8.952). Post hoc Tukey’s test: ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
(D) Regression Coefficient (k) summary for excitatory and inhibitory cohorts describing one sholl-based metric of neurite complexity. k values are negatively correlated to branching complexity. For excitatory versus inhibitory comparison: (unpaired t test ∗p < 0.05). For excitatory group: (one-way ANOVA: p < 0.0001, F[6,194] = 9.28). For inhibitory group: (one-way ANOVA: p < 0.0001, F[3,138] = 13.17). Post hoc Tukey’s test: ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
(E) Heatmap of changes in classifier accuracy for excitatory and inhibitory interneurons when metrics related to cell location, soma morphology, dendritic spines, or dendrite morphology are omitted from LDA (see STAR Methods for detailed metric membership in each category). Heatmap quantities are displayed as percent change in accuracy (true positive and true negative rate) when one of these categories are omitted, as compared to when all metrics are used to train the linear discriminant model.
(F) Percent quantification of action potential discharge patterns for excitatory (left) and inhibitory (right) cohorts. RF = Reluctant Firer, SS = single spiking, IB = Initial Bursting, p = Phasic, G = Gap, D = Delayed, RS = Regular Spiking; T = Tonic.
(G) Input Resistance for excitatory and inhibitory subtypes. For excitatory versus inhibitory cohort comparison: (unpaired t test ∗∗∗p < 0.0005). For excitatory group: (one-way ANOVA: p < 0.0001, F[6,70] = 9.516). For inhibitory group: (one-way ANOVA: p < 0.05, F[3,39] = 3.950). Post hoc Tukey’s test: ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
(H) Resting membrane potential for excitatory and inhibitory subtypes. For excitatory versus inhibitory cohort comparison: (unpaired t test: n.s.). For excitatory group: (one-way ANOVA: p < 0.001, F[6,10] = 5.966). For inhibitory group: (one-way ANOVA: p = 0.1918, F[3,39] = 1.658). Post hoc Tukey’s test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(I) Rheobase currents for excitatory and inhibitory subtypes. For excitatory versus inhibitory cohort comparison: (unpaired t test ∗∗∗∗p < 0.0001). For excitatory group: (one-way ANOVA: p = 0.0497, F[6,61] = 2.255). For inhibitory group: (one-way ANOVA: p = 0.9032, F[3,37] = 0.1891).