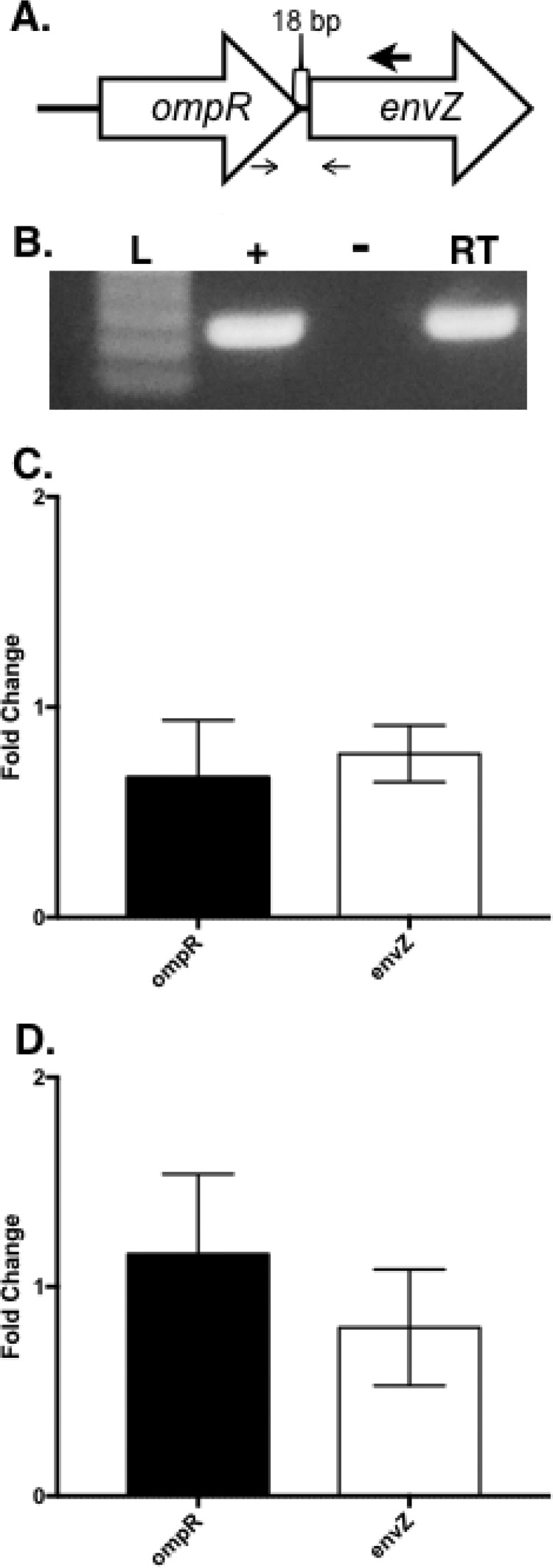
FIG 4.
The ompR-envZ operon is not autoregulated. (A) Graphic depiction of the organization of the ompR locus from A. baumannii strain AB5075. The large arrow above the envZ gene denotes the location of the primer utilized in the reverse transcriptase reaction to produce envZ-specific cDNA. The small arrows below ompR and envZ denote the locations of the primers spanning the 18-bp intergenic region between ompR and envZ. (B) Gel picture of PCR product from transcript-specific cDNA (generated from wild-type AB5075 RNA) used as the template with primers that span the intergenic region between ompR and envZ. Reactions are labeled “+” for positive control with genomic DNA, “−” for negative control RNA without reverse transcriptase, and “RT” for reverse transcriptase with envZ-specific cDNA as the template. (C) The expression of ompR and envZ was quantified by qRT-PCR on strains AB5075 and KT4 (ΔompR). Data are presented as the fold changes in expression ± the standard deviations for three replicates, as calculated by the Pfaffl method. (D) Expression of ompR and envZ was quantified by qRT-PCR on strain AB5075 grown with or without sodium chloride supplementation. Data are presented as fold changes in expression ± the standard deviations for three replicates, as calculated by the Pfaffl method.