Abstract
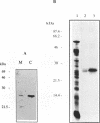
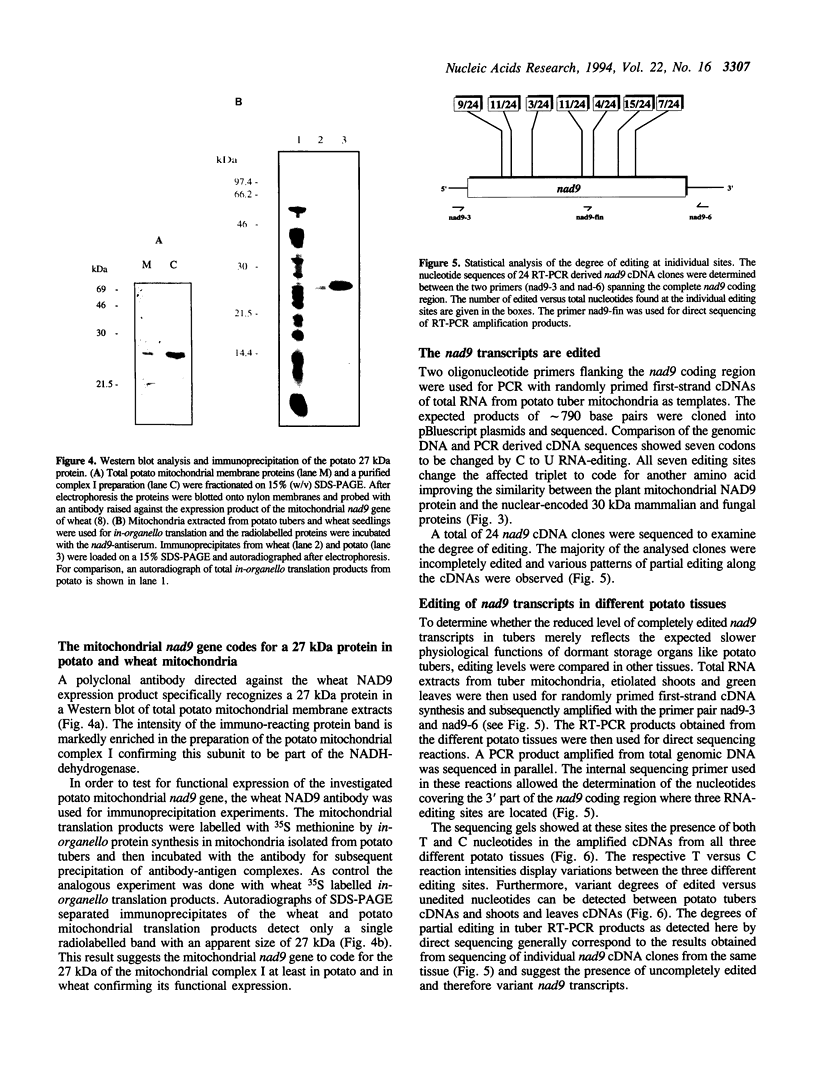
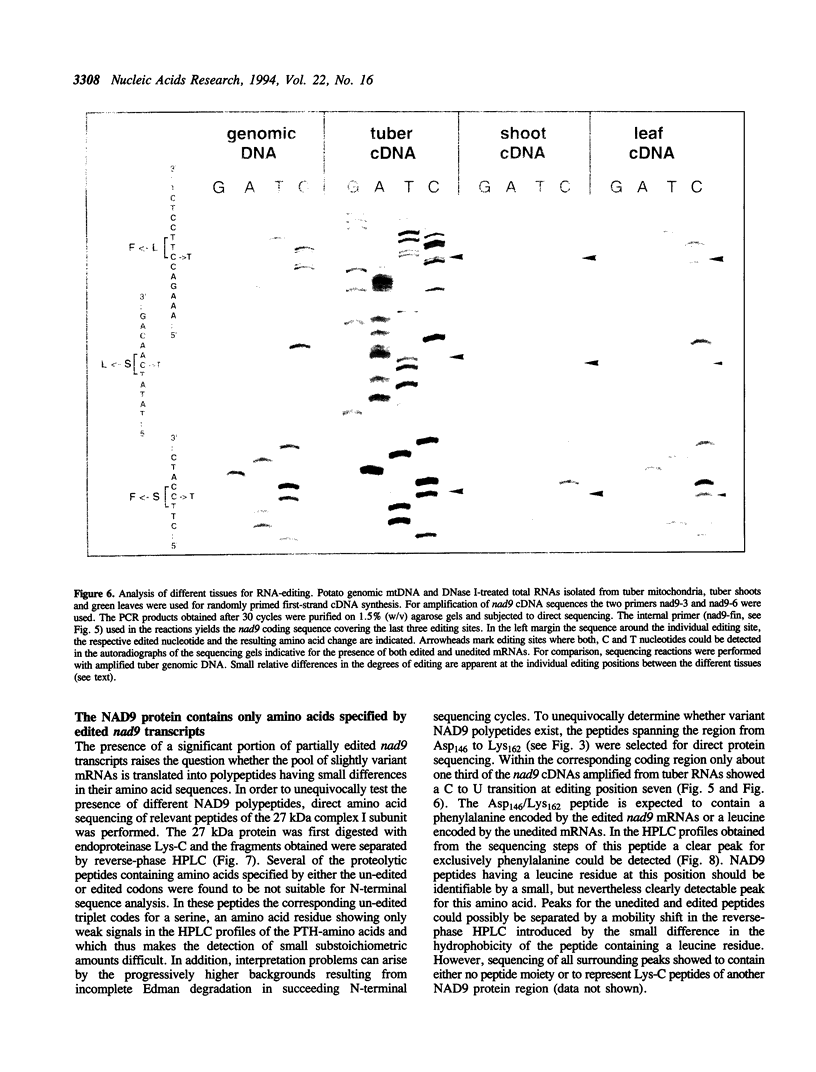
The pool of partially and completely edited mRNAs present in plant mitochondria could potentially be translated into a mixture of divergent proteins. This possibility was investigated for the nad9 gene in potato by characterization of the mRNA population and the corresponding protein sequence. The deduced amino acid sequence of the nad9 gene product has significant similarity to the nuclear-encoded 30 kDa subunit of the bovine and Neurospora NADH:ubiquinone oxidoreductase (complex I) and to the chloroplast ndhJ gene product. Immunoprecipitation of a 27 kDa in-organello 35S labelled mitochondrial translation product with an antibody directed against the wheat nad9 gene product demonstrates its functional expression in potato and wheat. Comparison of the nad9 genomic DNA and cDNA sequences reveals seven codons to be changed by a C to U RNA-editing. Direct sequencing of RT-PCR products derived from cDNAs of different tissues of potato plants shows the presence of a significant portion of only partially edited nad9 transcripts in the various tissues. Amino acid sequencing of internal peptides of the isolated 27 kDa protein from potato tubers demonstrates homogenous translation products of only completely edited nad9 mRNAs even in the presence of partially edited mRNAs. This result suggests a pretranslational selection between edited and incompletely edited mRNAs in plant mitochondria.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B., Lüke W., Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990 Mar 11;18(5):1309–1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., Hagemann R., Kössel H., Kudla J. Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids--a new regulatory mechanism? Mol Gen Genet. 1993 Aug;240(2):238–244. doi: 10.1007/BF00277062. [DOI] [PubMed] [Google Scholar]
- Bonen L., Williams K., Bird S., Wood C. The NADH dehydrogenase subunit 7 gene is interrupted by four group II introns in the wheat mitochondrial genome. Mol Gen Genet. 1994 Jul 8;244(1):81–89. doi: 10.1007/BF00280190. [DOI] [PubMed] [Google Scholar]
- Bégu D., Graves P. V., Domec C., Arselin G., Litvak S., Araya A. RNA editing of wheat mitochondrial ATP synthase subunit 9: direct protein and cDNA sequencing. Plant Cell. 1990 Dec;2(12):1283–1290. doi: 10.1105/tpc.2.12.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res. 1990 Sep 11;18(17):5189–5196. doi: 10.1093/nar/18.17.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T., Hofhaus G., Ise W., Nehls U., Schmitz B., Weiss H. A small isoform of NADH:ubiquinone oxidoreductase (complex I) without mitochondrially encoded subunits is made in chloramphenicol-treated Neurospora crassa. Eur J Biochem. 1989 Mar 1;180(1):173–180. doi: 10.1111/j.1432-1033.1989.tb14629.x. [DOI] [PubMed] [Google Scholar]
- Friedrich T., Weidner U., Nehls U., Fecke W., Schneider R., Weiss H. Attempts to define distinct parts of NADH:ubiquinone oxidoreductase (complex I). J Bioenerg Biomembr. 1993 Aug;25(4):331–337. doi: 10.1007/BF00762458. [DOI] [PubMed] [Google Scholar]
- Graack H. R., Grohmann L., Kitakawa M. The nuclear coded mitoribosomal proteins YmL27 and YmL31 are both essential for mitochondrial function in yeast. Biochimie. 1991 Jun;73(6):837–844. doi: 10.1016/0300-9084(91)90063-7. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Bonnard G., Lamattina L., Grienenberger J. M. Expression of the wheat mitochondrial nad3-rps12 transcription unit: correlation between editing and mRNA maturation. Plant Cell. 1991 Oct;3(10):1109–1120. doi: 10.1105/tpc.3.10.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäbler L., Herz U., Liddell A., Leaver C. J., Schröder W., Brennicke A., Grohmann L. The 42.5 kDa subunit of the NADH: ubiquinone oxidoreductase (complex I) in higher plants is encoded by the mitochondrial nad7 gene. Mol Gen Genet. 1994 Jul 8;244(1):33–40. doi: 10.1007/BF00280184. [DOI] [PubMed] [Google Scholar]
- Hernould M., Suharsono S., Litvak S., Araya A., Mouras A. Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2370–2374. doi: 10.1073/pnas.90.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz U., Schröder W., Liddell A., Leaver C. J., Brennicke A., Grohmann L. Purification of the NADH:ubiquinone oxidoreductase (complex I) of the respiratory chain from the inner mitochondrial membrane of Solanum tuberosum. J Biol Chem. 1994 Jan 21;269(3):2263–2269. [PubMed] [Google Scholar]
- Iwabuchi M., Kyozuka J., Shimamoto K. Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male sterile rice. EMBO J. 1993 Apr;12(4):1437–1446. doi: 10.1002/j.1460-2075.1993.tb05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky D. J., Riley G. R., Feagin J. E., Stuart K. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol Cell Biol. 1992 May;12(5):2043–2049. doi: 10.1128/mcb.12.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Mikami T., Kinoshita T. The sugar beet mitochondrial genome contains an ORF sharing sequence homology with the gene for the 30 kDa subunit of bovine mitochondrial complex I. Mol Gen Genet. 1993 Nov;241(3-4):479–481. doi: 10.1007/BF00284703. [DOI] [PubMed] [Google Scholar]
- Lamattina L., Gonzalez D., Gualberto J., Grienenberger J. M. Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem. 1993 Nov 1;217(3):831–838. doi: 10.1111/j.1432-1033.1993.tb18311.x. [DOI] [PubMed] [Google Scholar]
- Leterme S., Boutry M. Purification and preliminary characterization of mitochondrial complex I (NADH: ubiquinone reductase) from broad bean (Vicia faba L.). Plant Physiol. 1993 Jun;102(2):435–443. doi: 10.1104/pp.102.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monéger F., Smart C. J., Leaver C. J. Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J. 1994 Jan 1;13(1):8–17. doi: 10.1002/j.1460-2075.1994.tb06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K., Yamato K., Ohta E., Nakamura Y., Takemura M., Nozato N., Akashi K., Kanegae T., Ogura Y., Kohchi T. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992 Jan 5;223(1):1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- Pilkington S. J., Skehel J. M., Walker J. E. The 30-kilodalton subunit of bovine mitochondrial complex I is homologous to a protein coded in chloroplast DNA. Biochemistry. 1991 Feb 19;30(7):1901–1908. doi: 10.1021/bi00221a024. [DOI] [PubMed] [Google Scholar]
- Schuster W., Wissinger B., Unseld M., Brennicke A. Transcripts of the NADH-dehydrogenase subunit 3 gene are differentially edited in Oenothera mitochondria. EMBO J. 1990 Jan;9(1):263–269. doi: 10.1002/j.1460-2075.1990.tb08104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza A. E., Myler P. J., Stuart K. Maxicircle CR1 transcripts of Trypanosoma brucei are edited and developmentally regulated and encode a putative iron-sulfur protein homologous to an NADH dehydrogenase subunit. Mol Cell Biol. 1992 May;12(5):2100–2107. doi: 10.1128/mcb.12.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira A., Tropschug M., Werner S. Primary structure and expression of a nuclear-coded subunit of complex I homologous to proteins specified by the chloroplast genome. Biochem Biophys Res Commun. 1990 Sep 28;171(3):1168–1174. doi: 10.1016/0006-291x(90)90807-y. [DOI] [PubMed] [Google Scholar]
- Walker J. E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992 Aug;25(3):253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- Weidner U., Geier S., Ptock A., Friedrich T., Leif H., Weiss H. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993 Sep 5;233(1):109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- Yagi T., Xu X., Matsuno-Yagi A. The energy-transducing NADH-quinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Biochim Biophys Acta. 1992 Jul 17;1101(2):181–183. [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]