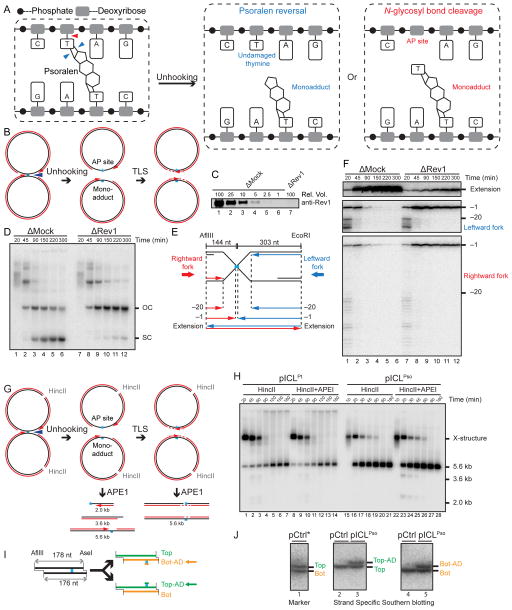
Figure 2. Psoralen-ICL is unhooked via the cleavage of a DNA N-glycosyl bond.
(A) Two possible incision-independent unhooking pathways for a psoralen-ICL. Blue arrowheads (left panel) show unhooking of a psoralen-ICL via breakage of the cyclobutane ring (psoralen reversal), resulting in an undamaged thymine and a monoadduct (middle panel). The red arrowhead (left panel) shows psoralen-ICL unhooking via cleavage of a DNA N-glycosyl bond, resulting in an AP site and a monoadduct (right panel).
(B) Model for pICLPso unhooking via cleavage of a DNA N-glycosyl bond, which necessitates gap filling of both daughter molecules by TLS.
(C) Mock-depleted and Rev1-depleted NPE were analyzed by Rev1 Western blotting. A relative volume of 100 corresponds to 0.25 μl NPE.
(D) pICLPso was replicated in mock or Rev1 depleted egg extracts in the presence of [α-32P]dATP. Repair intermediates were analyzed as in Figure 1C.
(E) Schematic illustration of nascent leading strands liberated after digestion of pICLPso with AflIII and EcoRI.
(F) pICLPso was replicated in mock- or Rev1-depleted egg extracts in the presence of [α-32P]dATP. Samples were purified and digested with AflIII and EcoRI before separation on a denaturing polyacrylamide gel. Three portions of the autoradiograph are shown with different contrasts for optimal display.
(G) Expected species after digestion of pICLPso replication intermediates with HincII and APE1, including 3.6 kb and 2.0 kb fragments.
(H) pICLPt or pICLPso was replicated in egg extracts with [α-32P]dATP. Replication intermediates were digested with HincII and APE1, separated on a native agarose gel, and visualized by autoradiography. Quantification of 3.6 kb and 2.0 kb fragments is presented in Figure S1J.
(I) Depiction of final repair products after AflIII and AseI digestion. AflIII and AseI generate different sized overhangs, allowing separation of top (178 nt) and bottom (176 nt) strands. Depending on which N-glycosyl bond is cleaved, the mono-adduct is present on the bottom (Bot-AD) or top (Top-AD) strands.
(J) Detection of pICLPso mono-adducts. pCtrl or pICLPso was replicated for 3 hours in egg extract. DNA was isolated, digested with AflIII and AseI, separated on a denaturing sequencing gel, and analyzed by strand-specific Southern blotting to visualize the top strand (middle panel) or bottom strand (right panel). The absence of bottom (lane 2) or top strands (lane 4) in the Southern blot of pCtrl indicates the strand specificity of the blotting protocol. To generate size markers for the top (178 nt) and bottom (176 nt) strands (left panel, lane 1), pCtrl was replicated in the presence of [α-32P]dATP (pCtrl*) and analyzed on the same sequencing gel after AflIII and AseI digestion.