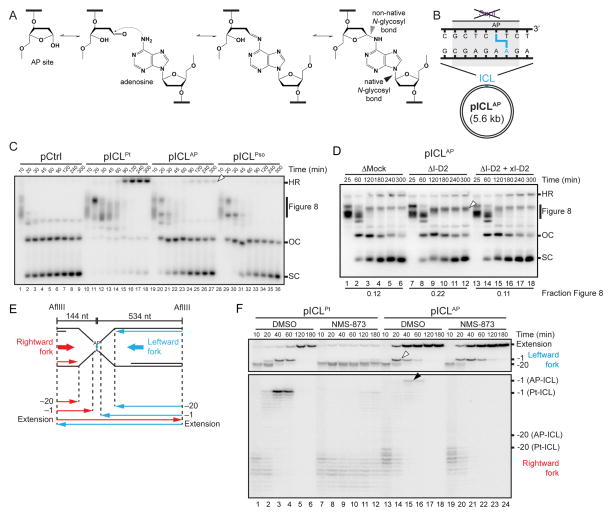
Figure 5. An AP-ICL is repaired by an incision-independent pathway that does not require FANCI-D2 or CMG unloading.
(A) Mechanism of AP-ICL formation.
(B) Cartoon of an AP-ICL containing plasmid (pICLAP), in which the SapI restriction site coincides with the ICL.
(C) pCtrl, pICLPt, pICLAP, or pICLPso was replicated in egg extracts and analyzed as in Figure 1C. White arrowhead, homologous recombination intermediate that accumulates for pICLAP.
(D) pICLAP was replicated in mock-depleted egg extract, FANCI-D2-depleted extract (ΔI-D2), or ΔI-D2 extract supplemented with xFANCI-D2 (ΔI-D2+xI-D2), and analyzed as in Figure 1C. White arrowhead, Figure 8 structures that persist in the absence of FANCI-D2. The fraction Figure 8 indicates the proportion of Figure 8 structures relative to total species at 300 min.
(E) Schematic illustration of nascent leading strands liberated by digestion of pICLAP with AflIII.
(F) pICLPt and pICLAP were replicated with [α-32P]dATP in the presence or absence of NMS-873. The nascent strands were purified and digested with AflIII before separation on a denaturing polyacrylamide gel. Two portions of the autoradiograph are shown with different contrasts for optimal display. White arrowhead, leftward -1 products. Black arrowhead, rightward -1 products.