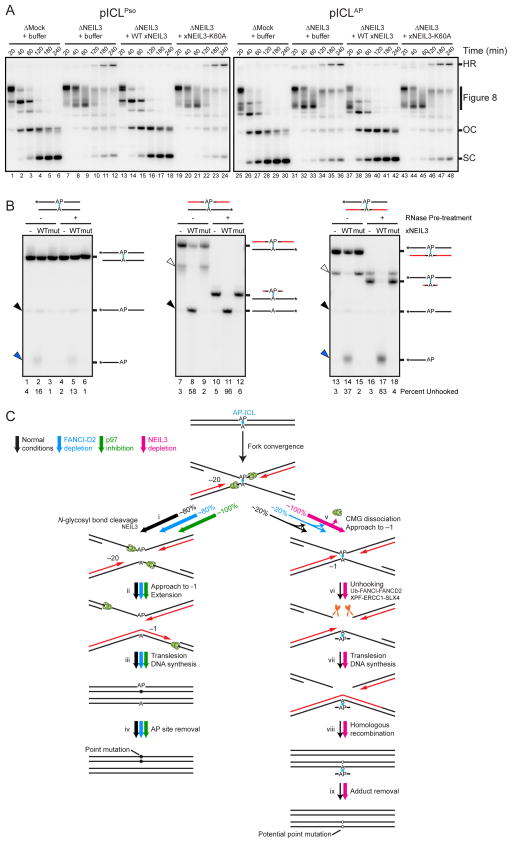
Figure 7. NEIL3 unhooks psoralen- and AP-ICLs.
(A) pICLPso or pICLAP was replicated in mock-depleted-, NEIL3-depleted-, or NEIL3-depleted egg extract supplemented with 300 nM recombinant NEIL3 in the presence of [α-32P]dATP. Repair intermediates were analyzed as in Figure 1C. As shown in Figure S7E, 15 nM recombinant NEIL3 also rescued pICLAP unhooking in NEIL3-depleted egg extract.
(B) Oligonucleotide substrates containing an AP-ICL were incubated with recombinant wild-type (WT) or K60A mutated (mut) NEIL3, separated on a denaturing polyacrylamide gel, and visualized by autoradiography. Black lines, DNA. Red lines, RNA. Cyan, AP-ICL. Asterisks indicate the 32P radiolabel. Black arrowheads, unhooked ssDNA. Blue arrowheads, β-elimination cleavage product. Bands marked by white arrowheads likely represent substrate that was not denatured during electrophoresis.
(C) Model for AP-ICL repair by incision-independent (left branch) or incision-dependent (right branch) pathways. Black lines, parental DNA. Red lines, nascent leading strands. Cyan, AP-ICL. Green ovals, CMG helicase. Pathway utilization is illustrated for unmodified extract (black arrows), FANCI-D2 depleted extract (blue arrows), extract supplemented with NMS-873 (green arrows), and NEIL3-depleted extract (pink arrows). Closed circles indicate point mutations that arise due to bypass of an AP site. Open circles indicate potential point mutations that may be introduced due to bypass of an AP-ICL-derived adenine monoadduct.