Fig. 2.
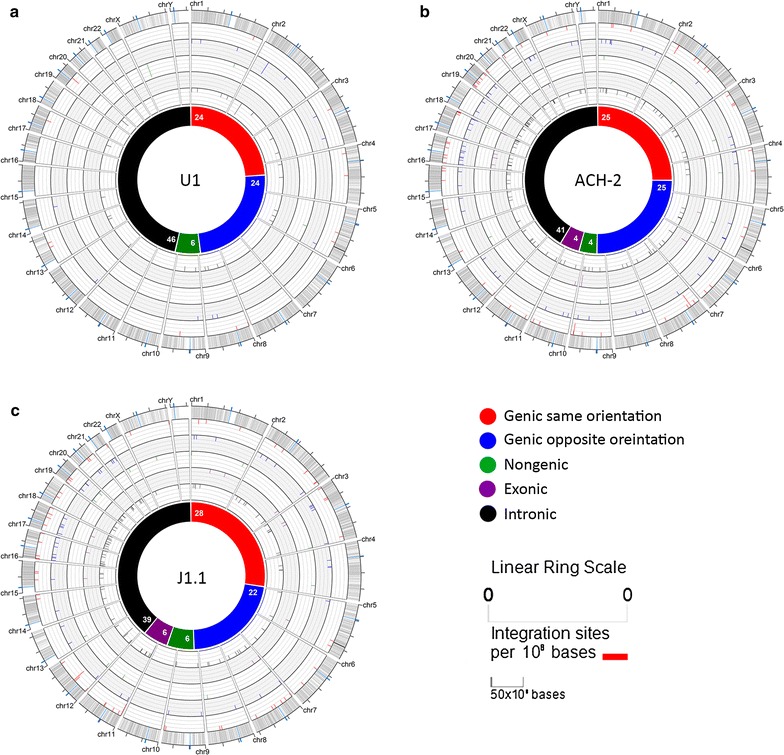
Site, frequency and nature of HIV integration sites in latently infected cells lines. A logarithmic depiction of HIV integration sites per million bases prior to passaging is shown for a U1, b ACH-2, and c J1.1. Chromosome numbers are labelled on the perimeter of the outer circle. The outer circle represents the different chromosomes with gene dense regions in grey bars representing different genes and in blue the centromere. The chromosomes are scaled to 50 million basepair bins. The integration sites for each chromosome are cumulative per million bases of the chromosome. The integration sites are shown as a coloured lines pointing to the centre of the circle. The frequency is represented in a logarithmic scale. The inner multi-colored solid circle summarises the nature of all the integration sites as genic (same or opposite orientation), non-genic, exonic and intronic. The specific site of integration is shown for each chromosome with the first ring showing integration within a gene with the same orientation compared to the host gene [genic (same orientation)] in red. The second ring shows HIV integration sites and within a gene with the opposite orientation compared to the host gene [genic (opposite orientation)] in blue. The third ring shows HIV integration sites not in a gene (non-genic) in green. In the fourth ring HIV integration sites in an exon (exonic) are in purple. In the fifth ring HIV integration sites in an intron (intronic) are in black
