Fig. 5.
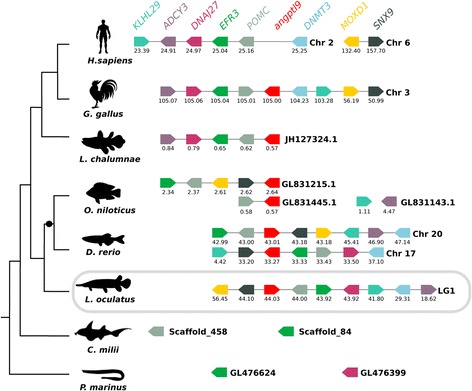
Comparison of the spotted gar genome region harbouring angptl9 with other fish, chicken and human. The gene environment of the spotted gar angptl9 gene was characterised and was used to identify homologous genome regions in the teleost (zebrafish and tilapia), the coelacanth (lobe-finned fish), the elephant shark (cartilaginous fish), marine lamprey (jawless fish), chicken and human. Horizontal lines represent the chromosome fragments; arrow boxes indicate genes and the arrowhead points to the orientation of the predicted gene transcription. Only genes that were conserved across species are represented. Gene symbols are indicated and homologue genes are represented by the same colour and they are represented according to their order in the chromosome. The size of the genome fragments analysed and the location of the gene in the chromosome are indicated in Megabase pairs. “●”: Teleost Specific Genome Duplication (TSGD). Gene names and symbols are: Kelch-like family member 29 (KLHL29), Adenylate cyclase 3 (ADCY3), DnaJ (HSP40) homolog, subfamily C member 27 (DNAJC27), EFR3 (EFR3), Proopiomelanocortin (POMC), DNA (cytosine-5)-methyltransferase 3 (DNMT3), Monooxygenase, DBH-like 1 (MOXD1) and Sorting nexin 9 (SNX9)
