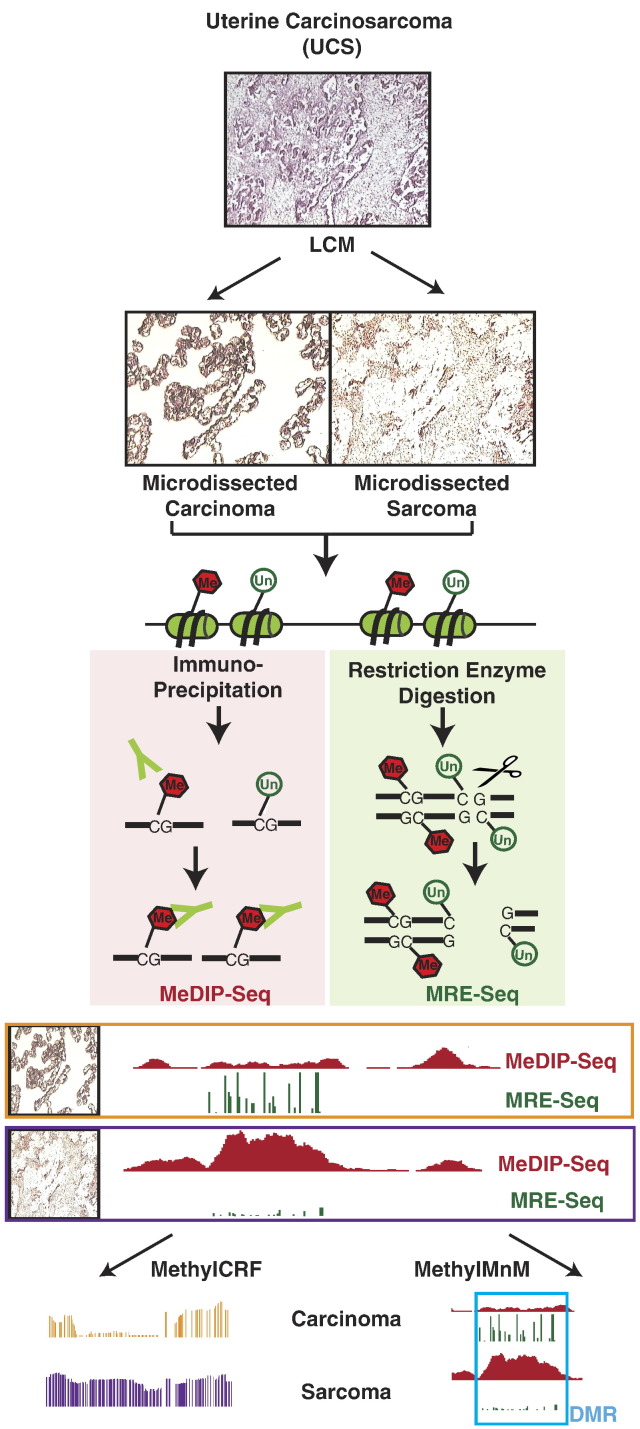
Fig. 1.
Basic workflow for DNA methylome sequencing of LCM samples. After LCM, distinct carcinoma components and sarcoma components dissected from UCS were sequenced using MeDIP-seq and MRE-seq. MeDIP-seq: After sonication, the methylated genomic DNA fragments were captured by a monoclonal anti–5′-methylcytosine antibody. Immunoprecipitated DNA fragments were analyzed by high-throughput DNA sequencing. MRE-seq: Instead of sonication, the genomic DNA was digested by multiple methylation-sensitive restriction enzymes. Specific size fragments were selected and then sequenced. The bioinformatics pipeline combined two algorithms (methylCRF and M&M) to estimate absolute methylation levels at single-CpG resolution and to detect DMRs between the two components, respectively.