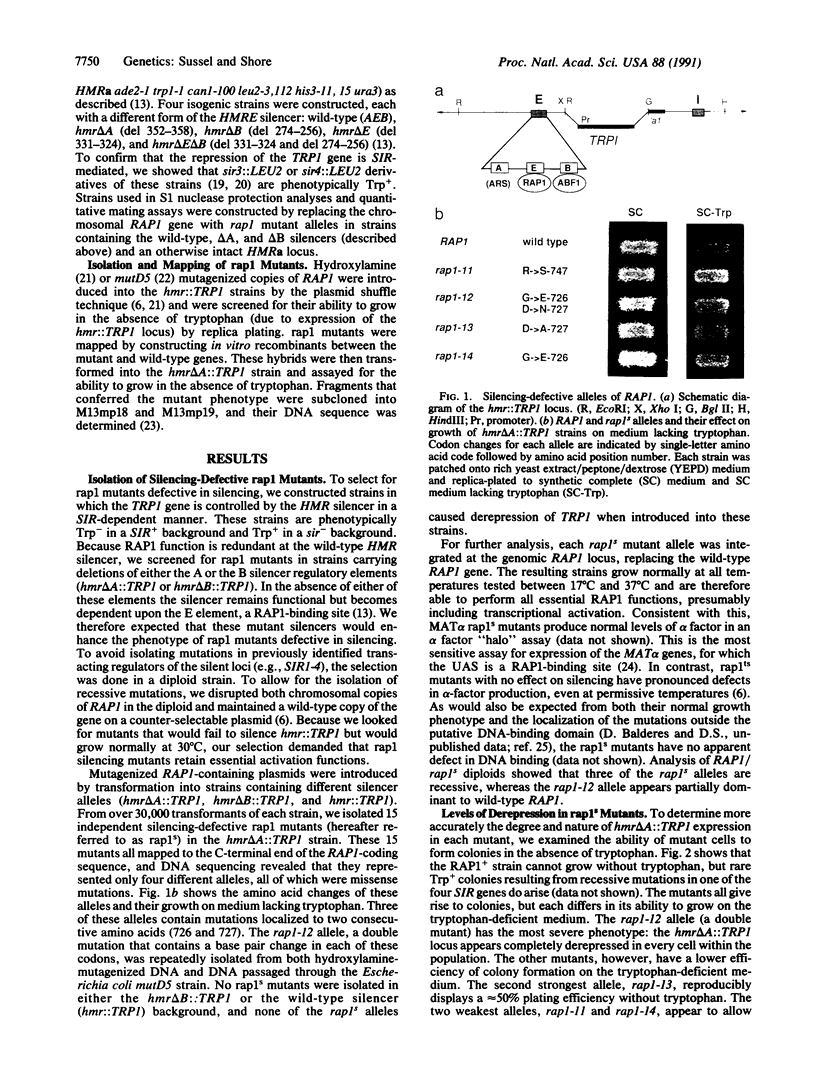
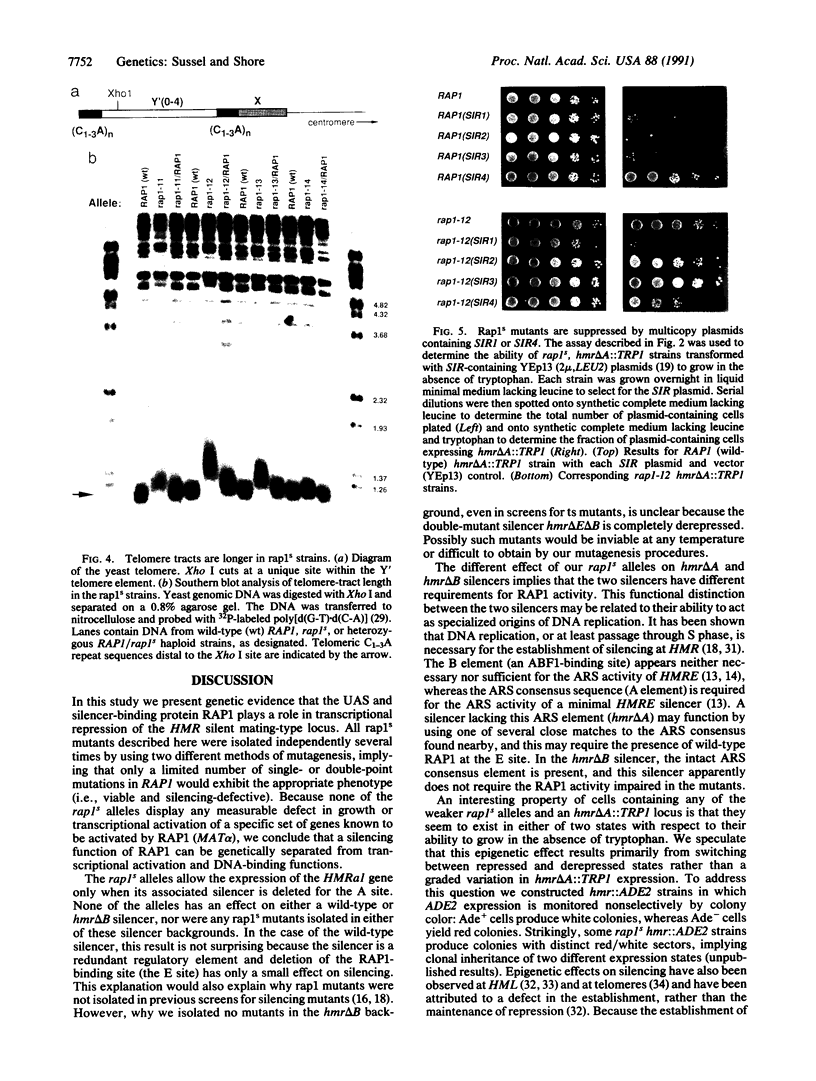
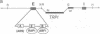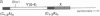Abstract
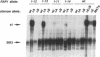
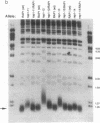
The repressor/activator protein 1 (RAP1) binds to the upstream activating sites of many genes, the silencer elements flanking the unexpressed mating-type loci HMR and HML, and the poly(C1-3A) sequences at telomeres, suggesting that RAP1 might have three distinct regulatory functions. To determine the in vivo role of RAP1 in repression of the HMR silent locus, we developed a screen to isolate rap1 mutants specifically defective in silencing. Fifteen independent mutants defining four different rap1 alleles were isolated. These alleles are defective to different extents in repression of an HMR locus containing a mutated, but fully functional, silencer. All four alleles are missense mutations in only three codons within a small C-terminal region of the gene. These silencing-defective mutants have no apparent growth defects, indicating that expression of the large number of essential genes that have promoters containing RAP1-binding sites is normal. A transcriptional silencing function of RAP1 can therefore be genetically separated from its presumably essential activation functions. Surprisingly, three of the silencing-defective rap1 alleles have significantly longer telomeres, suggesting that the function of RAP1 in both transcriptional silencing and telomere-length regulation may be related. In addition, we have demonstrated that increased gene dosage of either SIR1 or SIR4, two other factors required for silencing, suppresses the silencing defect of the rap1 mutants. The properties of SIR4 dosage suppression suggest that SIR4 protein may interact directly with RAP1 at silencers.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bankier A. T., Weston K. M., Barrell B. G. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 1987;155:51–93. doi: 10.1016/0076-6879(87)55009-1. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990 Nov 16;63(4):739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974 Feb;117(2):477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Shenk T. Adenovirus E1A protein paradigm viral transactivator. Annu Rev Genet. 1989;23:141–161. doi: 10.1146/annurev.ge.23.120189.001041. [DOI] [PubMed] [Google Scholar]
- Giesman D., Best L., Tatchell K. The role of RAP1 in the regulation of the MAT alpha locus. Mol Cell Biol. 1991 Feb;11(2):1069–1079. doi: 10.1128/mcb.11.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Henry Y. A., Chambers A., Tsang J. S., Kingsman A. J., Kingsman S. M. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990 May 11;18(9):2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991 Apr;5(4):616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Wilson N. M., Petracek M. E., Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989 Oct;16(4):225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- Lustig A. J., Kurtz S., Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990 Oct 26;250(4980):549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- Mahoney D. J., Marquardt R., Shei G. J., Rose A. B., Broach J. R. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991 Apr;5(4):605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney D., Rose A., Hicks J. B., Broach J. R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. M., Nasmyth K. A. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984 Nov 15;312(5991):247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Miller A. M., Sternglanz R., Nasmyth K. A. The role of DNA replication in the repression of the yeast mating-type silent loci. Cold Spring Harb Symp Quant Biol. 1984;49:105–113. doi: 10.1101/sqb.1984.049.01.014. [DOI] [PubMed] [Google Scholar]
- Miller A. M. The yeast MATa1 gene contains two introns. EMBO J. 1984 May;3(5):1061–1065. doi: 10.1002/j.1460-2075.1984.tb01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Pillus L., Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989 Nov 17;59(4):637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Rine J., Strathern J. N., Hicks J. B., Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979 Dec;93(4):877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D. D., Helms S., Carlstedt-Duke J., Gustafsson J. A., Rottman F. M., Yamamoto K. R. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988 Sep;2(9):1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- Shampay J., Blackburn E. H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Jan;85(2):534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. DNA sequences of telomeres maintained in yeast. Nature. 1984 Jul 12;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Squire M., Nasmyth K. A. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984 Dec 1;3(12):2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Boeke J. D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Herskowitz I. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J Mol Biol. 1981 Dec 5;153(2):305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Swanson M. J., Romeo A. M., Hicks J. B., Sternglanz R. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol Cell Biol. 1991 Apr;11(4):2253–2262. doi: 10.1128/mcb.11.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]