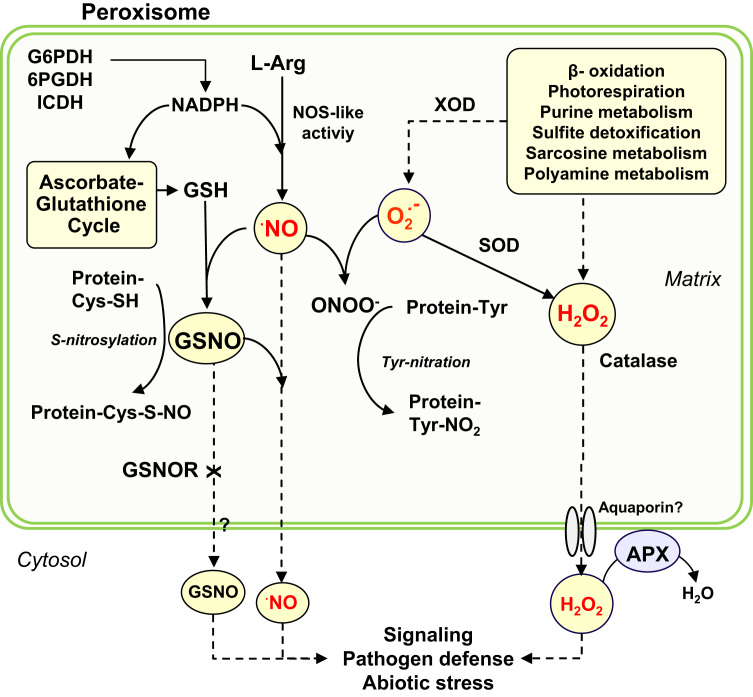
Fig. 2.
Model of NO and H2O2 metabolism in plant peroxisomes.L-Arginine nitric oxide synthase (NOS)-like activity generates NO which can react with reduced glutathione (GSH) in the presence of O2 to form S-nitrosoglutathione (GSNO). This metabolite can interact with SH-containing proteins by a process of S-nitrosylation affecting their function. NO can also react with superoxide radicals (O2·-) to generate peroxynitrate (ONOO-) which can mediate a process of tyrosine nitration of proteins. Alternatively, either NO or GSNO can be released to the cytosol to participate in signaling cascades, although the S-nitrosoglutathione reductase (GSNOR) could modulate the GSNO release. Hydrogen peroxide (H2O2) is produced by different biochemical pathways. Additionally, the superoxide radical (O2·-) generated by some enzymes such as xanthine oxidase (XOD) which is part of purine metabolism can dismutate into H2O2 by the enzyme superoxide dismutase (SOD). The level of peroxisomal H2O2 is controlled either by catalase located in the matrix or by the membrane bound ascorbate peroxidase (APX), and its release to the cytosol could be through putative aquaporins.