FIGURE 7.
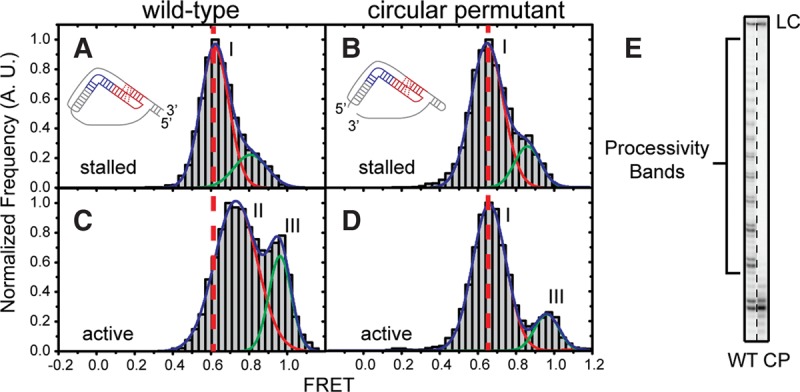
Template-pseudoknot fold connectivity is crucial for motion of the pseudoknot fold and telomerase processivity. (A,C) Single-molecule FRET histograms generated by a WT RNP containing a U92-U312 FRET pair before (A) and after (C) addition of activity buffer. The RNA cartoon depicts the 5′ and 3′ ends of the core domain used for telomerase reconstitution. The vertical dotted line delineates the observed change in FRET upon addition of activity buffer. (B,D) Single-molecule FRET histograms of RNPs containing a circular permutant RNA at hTR 62–63 RNP and a U92–U312 FRET pair, before (B) and after (D) addition of activity buffer. The vertical dotted line delineates the lack of FRET change observed upon addition of activity buffer. Roman numeral peak labels (i,ii, and iii) reference the different FRET peaks that give rise to the separate models in Figure 3. (E) Activity assay of WT and circular permutant enzymes with a (TG)6TTAGGG primer substrate. WT products show binding, extension, and translocation, resulting in processivity bands. Circular permutant complexes exhibit binding and extension, but failed translocation and therefore a lack of processivity bands.
