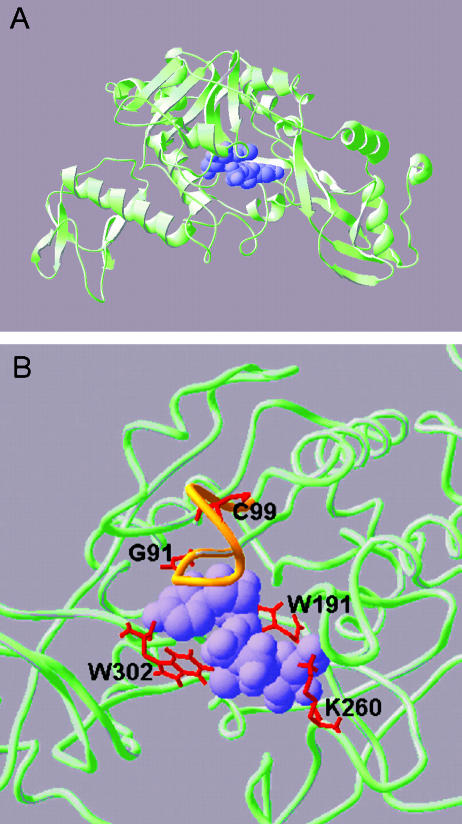
Figure 3.
Predicted three-dimensional structure of barley UGPase. The overall UGPase structure (A) was predicted using Swissmodel first approach method using human AGX-1 as a template fold and barley UGPase protein sequence. The model shows segments of UGPase as α-helices (curls), β-sheets (ribbons with arrows), and coils (wire tubes). The predicted location of the substrate (UDPG) is shown as a purple space-filled model. In the enlargement of the active site, shown in wireframe view (B), the nucleotide binding (NB) loop conserved in all pyrophosphorylases is colored orange. Residues shown by mutation to be critical to activity are shown in red and labeled in amino acid unicode (e.g. W = Trp, K = Lys) along with their position in the barley UGPase protein sequence. Illustrations were generated by Swissprotein Deepview (v 3.7).