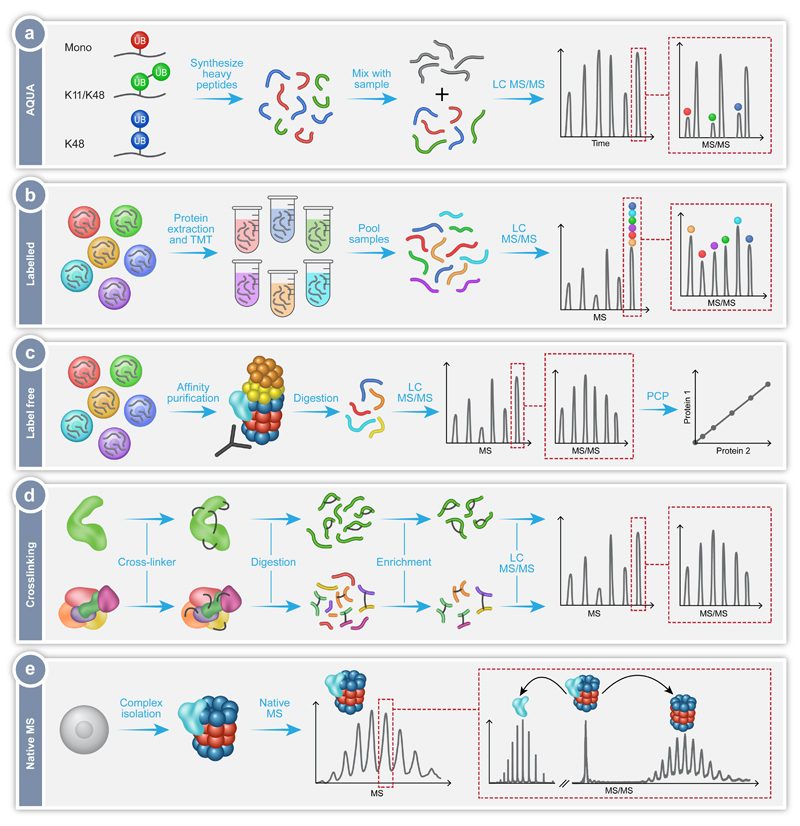
Fig. 2.
MS workflows utilized in the exploration of the proteasomal protein degradation pathway. (a) Absolute quantification of ubiquitination using the AQUA technique, in which synthetic heavy peptides corresponding to the types of Ub-linkages are produced, for example mono-Ub (red), heterogeneous poly-Ub containing K11 and K48 linkages (green), or homotypic poly-Ub containing only K48 linkages (blue). Known quantities of these peptides are then combined with the peptides from the sample under investigation (gray) and analyzed by LC-MS/MS. The synthetic heavy peptide and light analyte peptide will have the same retention time during chromatography, thus eluting in the same peak. The peaks can be individually selected and subject to MS/MS, with the mass difference between the light and heavy peptides now distinguishable. Consequently, the ratio between the native peptide and the isotope labeled AQUA standard peptide enables absolute quantification. (b) Labelling of protein extracts using Tandem Mass Tagging (TMT). Proteins are extracted from several cell lines under investigation, followed by the differential labelling of each extract with a unique isobaric tag. The samples are digested and the produced peptides from each sample pooled together. This mixture is then analyzed by LC-MS/MS with the same peptide from each sample falling under the same m/z peak. Selection of this peak followed by MS/MS leads to fragmentation and separation of the differently tagged peptides. The relative peptide levels from each sample can then be quantified and compared. (c) Label free quantification of isolated protein complexes. In this method the different samples are analyzed separately and compared. Often, the protein complex of interest is extracted from a cell lysate using affinity purification. The complex is then digested and subjected to LC-MS/MS allowing identification of the components of the complex and any modifications they contain. By measuring the intensity or ion counts of the precursor ions and comparing this value across samples, relative quantification is achieved. Protein correlation profiling (PCP) can then be performed to determine the composition of the protein complexes under investigation. (d) Crosslinking coupled MS of proteins (top) and protein complexes (bottom). The addition of crosslinkers to the proteins of interest will cause the formation of covalent bonds between the crosslinker and two amino acids (black lines). These can be within the same protein or between two different proteins. The crosslinked samples are then digested to produce peptides, some of which will contain the crosslink. Uncrosslinked peptides can be removed during an enrichment step. The crosslinked peptides are then subjected to LC-MS/MS, with specific peak selection and fragmentation allowing the identification of the location of the crosslink, and consequently the determination of the proximity of the amino acids involved. (e) Native MS of intact protein complexes. Whole protein complexes are isolated, purified and subjected to MS analysis. The intact mass of the complex reflects the homogeneity and stability of the sample as well as its subunit stoichiometry. Isolation of peaks followed by MS/MS fragmentation leads to the dissociation of distinct associating proteins or subunits, which can then be individually identified. This step also enables differentiation between core and peripheral subunits as well as revealing protein-protein connectivities.