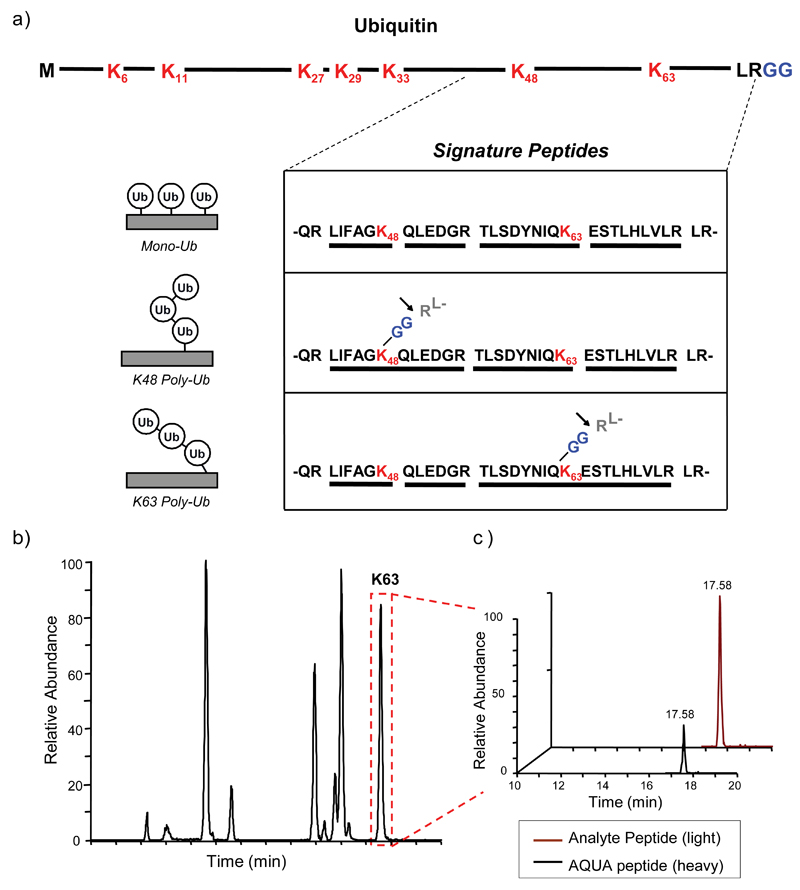
Fig. 3.
Quantification of Ub-linkages using AQUA and LC-MS/MS. (a) Ubiquitin contains seven lysine (K) residues in its primary sequence (highlighted in red) and a C-terminal diglycine (GG) motif (highlighted in blue). Tryptic digestion will cleave after K and R residues, producing unique peptides for mono-Ub (top), K48 poly-Ub (middle) and K63 poly-Ub (bottom) linkages, as well as after the R at the C-terminus of Ub (indicated by arrow), leading to the identifiable –GG motif on K48 and K63 linked peptides. These signature peptides are isotopically labelled and added to a sample at known concentrations, followed by analysis by LC-MS/MS. (b) A representative analysis of a peptide sample containing free K63-linked poly-Ub chains, showing the retention time of K63 linked Ub peptides (boxed in red) during LC-MS. Both the heavy signature peptides and light analyte peptides have the same retention time. (c) SRM (MS/MS) analysis of the K63 peak showing the mass difference between the heavy AQUA peptide and the light analyte peptide. Due to the known concentration of the AQUA peptide added to the sample, the amount of analyte peptide can be quantified. Reproduced and adapted with permission from (Kirkpatrick et al., 2006).