Figure 5. PE6 extends yeast CLS independently of presently known longevity-defining signaling pathways/protein kinases.
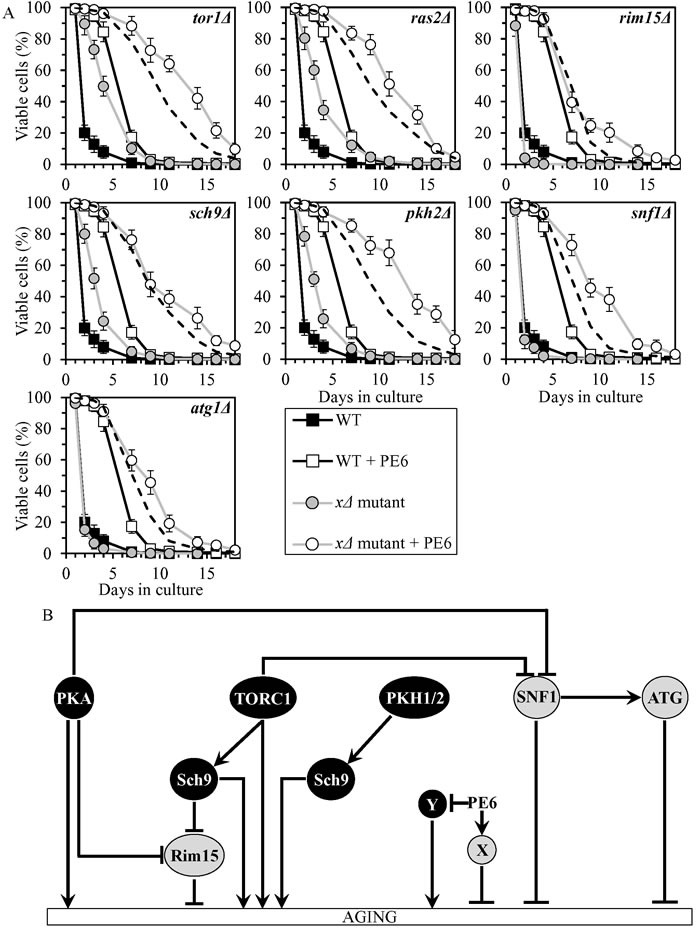
A. Cells of the wild-type (WT) and indicated mutant strains were grown in the synthetic minimal YNB medium (0.67% Yeast Nitrogen Base without amino acids) initially containing 2% glucose, in the presence of 1.0% PE6 (ethanol was used as a vehicle at the final concentration of 5.0%) or in its absence (cells were subjected to ethanol-mock treatment). Survival curves of chronologically aging WT and mutant strains cultured with or without 1.0% PE6 are shown. Data are presented as means ± SEM (n = 8). The dotted line indicates the predicted survival curve of a particular mutant strain cultured with PE6 if this PE exhibits an additive longevity-extending effect with the mutation. Data for the mock-treated WT strain and for the WT strain cultured with PE6 are replicated in all graphs of this Figure. B. The effects of PE6 on anti-aging and/or pro-aging processes that are not controlled by the network of presently known signaling pathways/protein kinases. These effects are inferred from the data presented in A., Tables 2 and 3, and Suppl. Figs. S3 and S9. Abbreviations: as in the legend to Figure 1.
