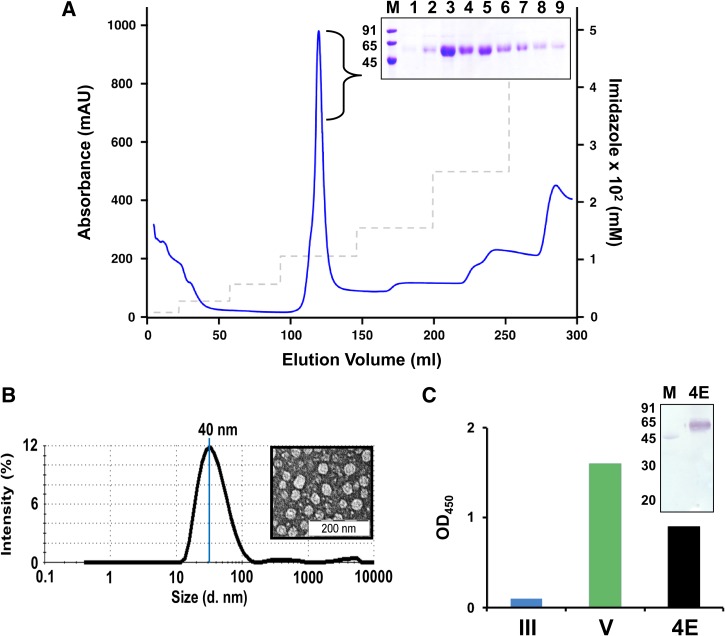
Figure 2.
Purification and characterization of DENV-4 E. (A) DENV-4 E was purified from pellet fraction of lysate of induced Pichia pastoris biomass via Ni2+ affinity chromatography. Blue curve and dashed grey line represent the UV-absorbance profile and imidazole step gradient, respectively. The inset represents Coomassie-stained SDS-PAGE analysis of the peak fractions (lanes 1–9) of purified DENV-4 E. Lane M represents low-molecular-weight protein markers, the sizes (in kDa) of which are written on the left of the inset. (B) Formation of virus-like particles by purified DENV-4 E as assessed by DLS representing particle size distribution by intensity. The average particle size is also indicated. EM image of the same is provided in the inset. (C) Glycosylation of DENV-4 E (4E, black bar) is evaluated by ELISA using Con A-HRPO conjugate on Ni-NTA His-Sorb plate. EDIII (III, blue bar) and DENV-4 (V, green bar) is used as negative control and positive control, respectively. In the inset, a protein blot using Con A-HRPO conjugate is shown in which lane M represents low-molecular-weight protein markers, which contain ovalbumin. Low-molecular-weight protein markers serve as positive (through its only glycosylated protein, ovalbumin) and negative (proteins other than ovalbumin) controls in the assay, and the sizes (in kDa) of the protein markers are indicated on the left. Con A-HRPO = Concanavalin A-horseradish peroxidase; DENV-4 E = dengue virus serotype 4 ectodomain; ED = envelope domain; ELISA = enzyme-linked immunosorbent assay.