Fig. 2.
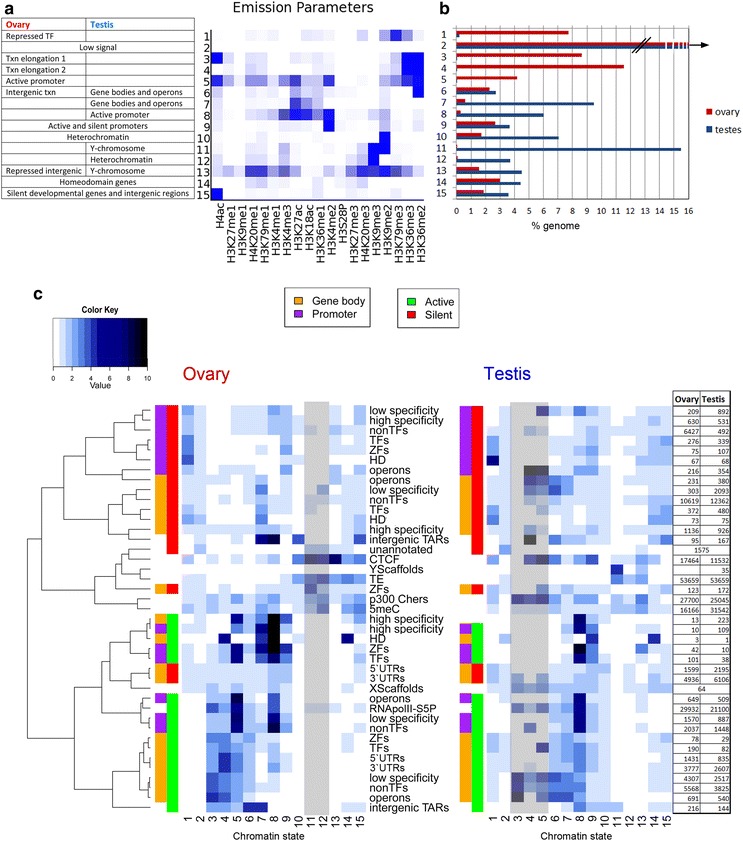
Chromatin states in the O. dioica ovary and testis reveal two distinct epigenetic landscapes. Heatmap of model emission parameters (a) for 15 chromatin states learned across the genome, using both samples (see also Additional file 3: Table S3). Proportions of the genome in the ovary and testis covered by each chromatin state (b) show sex/tissue specificity of chromatin states. Heatmaps (c) visualize the fold enrichments of each chromatin state (columns) for a set of genomic features (rows), as listed, in the ovary and testis. This facilitates assignment of putative biological function(s) of each chromatin state in each tissue. Gray shading indicates states that cover below 0.1% of the genome (70,000 bp) in each tissue. These low-coverage states nevertheless have large fold enrichments over certain genomic features, e.g., state 3 has low coverage in the testis but is found more often than expected by chance at active operons. Features are clustered according to their correlation in the ovary, and this ordering is used for the testis heatmap to allow comparison. The table gives the numbers of each feature in each sample. Gene bodies (orange) and promoter regions (purple) are split into active (green) and silent (red) states in the respective ovary and testis tissues. Txn, transcription; TF, transcription factor; ZF, zinc finger protein; HD, homeodomain protein, high- and low-specificity genes according to breadth of expression across development (see “Methods” section); TAR, transcriptionally active region; TE, transposable element; unannotated, regions lacking annotation or transcription; 5meC, methylcytosine; RNApolII-S5P, serine 5 phosphorylated RNA polymerase II
