Fig. 5.
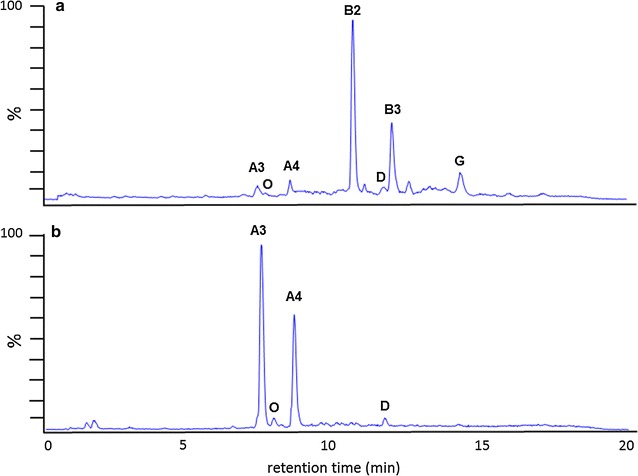
UPLC-qTOF-HR-MS/MS analysis of the extracts obtained from the mutant strains. a UPLC-qTOF-MS/MS chromatogram selected for m/z = 527.3014 corresponding to milbemycin A3 (A3), m/z = 541.3171 corresponding to milbemycin A4 (A4) and milbemycin B2 (B2), m/z = 555.3327 corresponding to milbemycin D (D) and milbemycin B3 (B3), m/z = 569.3484 corresponding to milbemycin G (G), and m/z = 789.5158 corresponding to oligomycin A (O), respectively, of culture extracts from the S. avermitilis SAMA13 and SAMA1M7 strains. b UPLC-qTOF-MS/MS chromatogram selected for m/z = 527.3014, 541.3171, 555.3327, and 789.5158 corresponding to milbemycin A3 (A3), milbemycin A4 (A4), milbemycin D (D), and oligomycin A (O), respectively, of culture extracts from the S. avermitilis SAMA1M7ΔD strain
