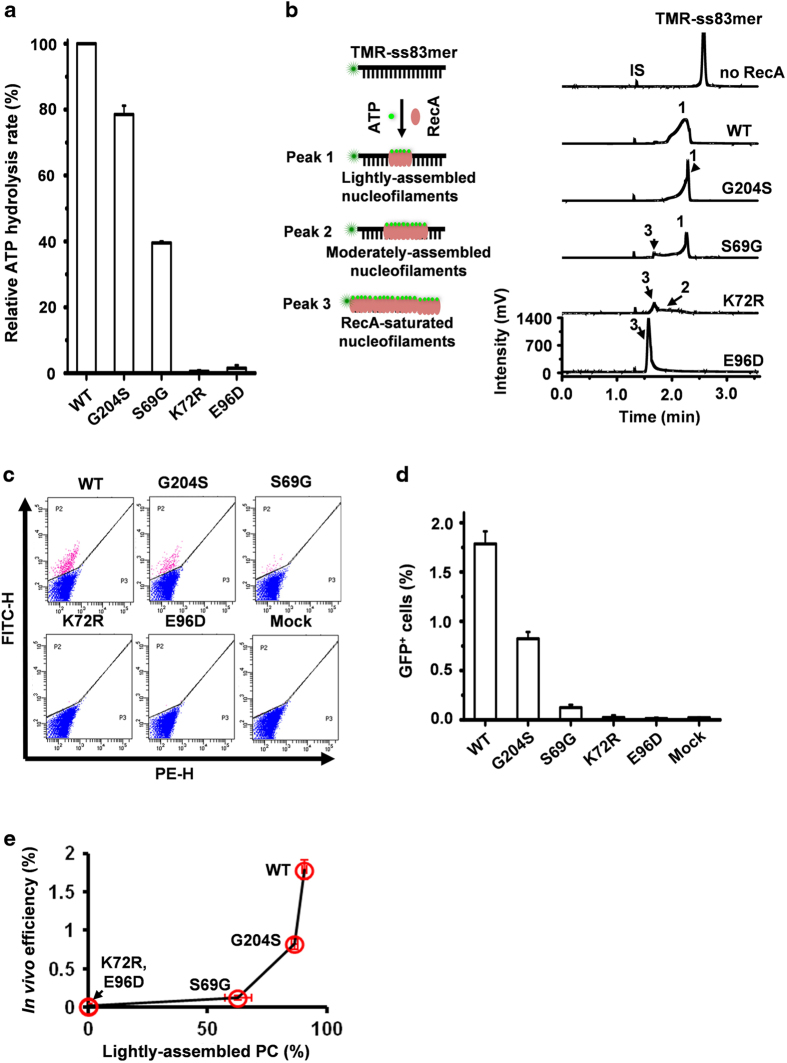
Figure 5.
In vivo E. coli homologous recombination repair and its correlation with in vitro lightly assembled nucleofilaments. (a) The ATP hydrolytic activity of four representative RecA mutants (G204S, S69G, E96D and K72R). The activity was normalized to that of the wild type of RecA. The reactions contained 3.0 μM RecA, 10 nM ss83mer and 1.0 mM ATP. (b) Electropherograms from CE-LIFP analysis of the mutated RecA-ssDNA filaments. The reactions contained 10 nM TMR-ss83mer, 3.0 μM RecA and 1.0 mM ATP, and proceeded at 37 °C for 10 min. IS indicates internal migration marker. Peaks 1–3 represent the lightly assembled, moderately assembled and saturated nucleofilaments, respectively. (c, d) FACS analysis of E. coli HR efficiency of four RecA mutants (G204S, S69G, E96D and K72R) (c) and the corresponding percentage of GFP+ cells (d). FITC-H and PE-H (c) indicate the GFP signal and background fluorescence of the sorted E. coli cells, respectively. FITC: ex 488 nm, em 530 nm; and PE: ex 488 nm, em 585 nm. Pink and blue dots (c) indicate GFP+ and GFP− cells, respectively. (e) The correlation of in vivo HR efficiency (ref. to d) with in vitro lightly assembled nucleofilaments as indicated in b.