Figure 7.
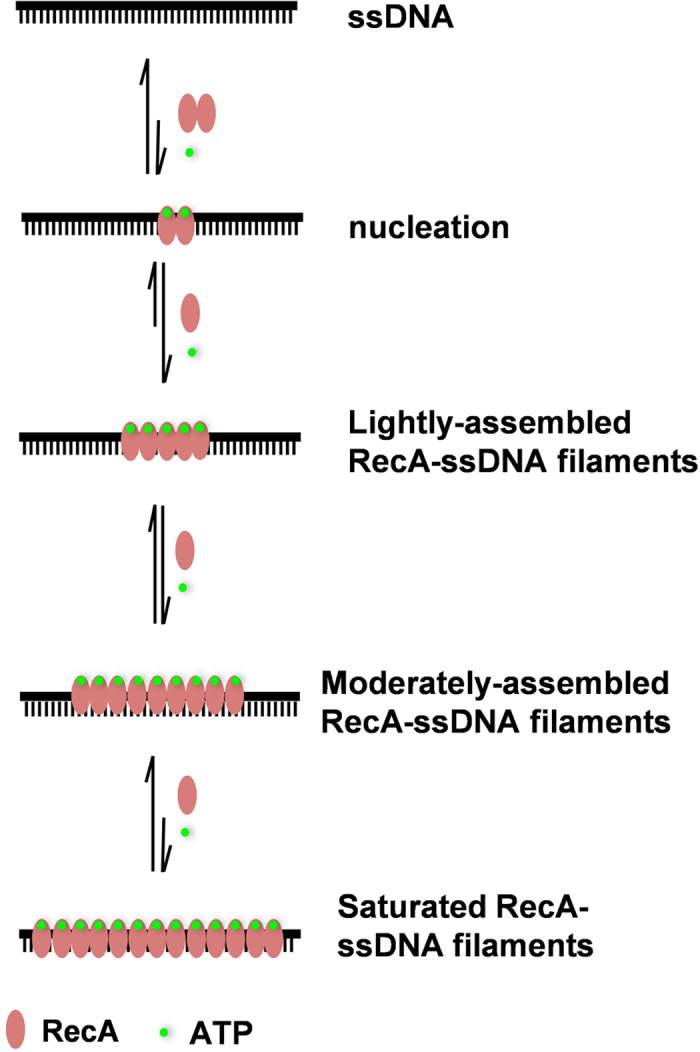
Dynamic model for the formation of diverse RecA nucleofilaments as stimulated by physiologically relevant ATP. First, two RecA monomers cooperatively nucleate on ssDNA; second, the nucleated RecA dimer further extends by assembling one RecA monomer on ssDNA each step to form the lightly assembled filaments; third, by same mechanism extends to form the moderately assembled filaments; and fourth, finally extends to form the saturated filaments. Due to tight regulation of the filaments by the RecA ATPase hydrolytic activity, the longer filaments will have more ATP hydrolysis and cause significant RecA dissociation, thereby, the unsaturated filaments (the lightly assembled filaments and the moderately assembled filaments) predominate and the saturated filaments are minor.
