Figure 4.
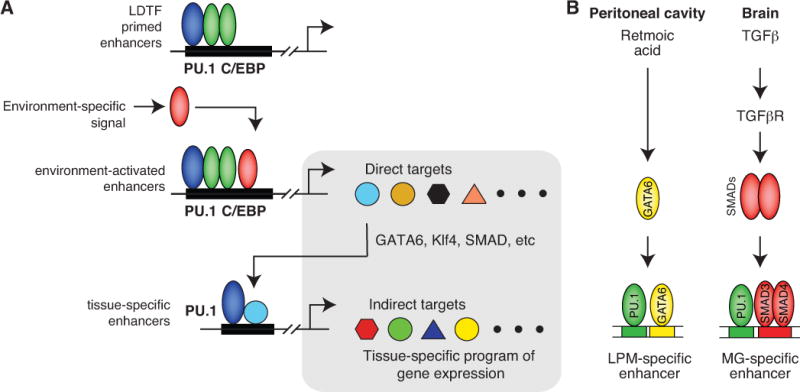
Selection and activation of tissue-specific macrophage enhancers. A. Generic model. A core set of macrophage LDTFs, exemplified by PU.1 and C/EBP factors, prime a common set of enhancers in many or all macrophage subsets. These enhancers can be acted upon by environment-specific signals to drive the expression of direct target genes. A subset of these genes includes transcription factors that can collaborate with macrophage LDTFs, such as PU.1, to select a secondary, tissue-specific set of enhancers that drive expression of additional target genes. The tissue-specific gene expression program thus results from both direct and indirect environmental effects. B. Examples of signals preferential for the peritoneal cavity (retinoic acid) or brain (TGFβ), resulting in expression of collaborative factors Gata6 or Smads, respectively.
