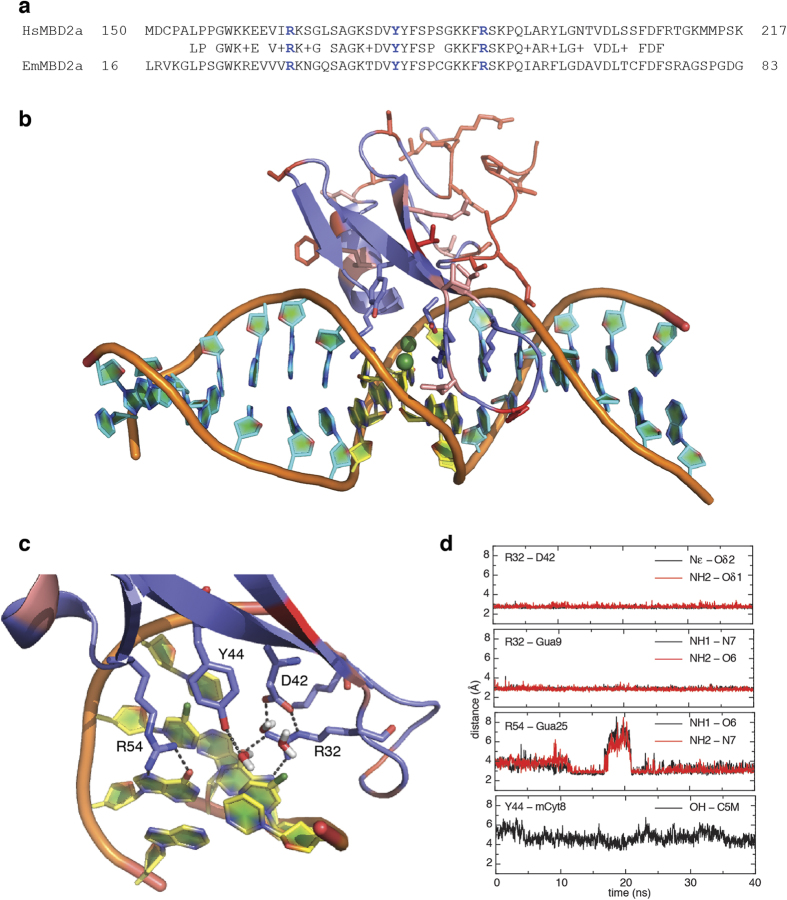
Figure 2. Structural conservation of the MBD of MBD2.
(a) Alignment of the MBDs from EmMBD2/3 and HsMBD2 show a high degree of identity including residues critical for recognizing methylated cytosines (blue). (b) A cartoon diagram is shown of the final frame from a 40 ns molecular dynamics simulation of the MBD from EmMBD2. Residues that differ between HsMBD2 and EmMBD2/3 are shown as sticks in pink (conservative changes) and red (non-conservative changes) and residues critical for recognizing mCpG are shown as sticks in blue. The mCpG dinucleotide is shown in yellow with the methyl group highlighted as a green sphere. A close-up view (c) shows that the critical residues at the protein-DNA interface form the appropriate structure and hydrogen bonding network for methylation selectivity including several water molecules surrounding the methyl group of methyl-cytosine. (d) The distances between key hydrogen bond donor and acceptors and between the tyrosine hydroxyl and methyl carbon are plotted. These critical interactions are highly stable and maintained throughout the duration of the simulation.