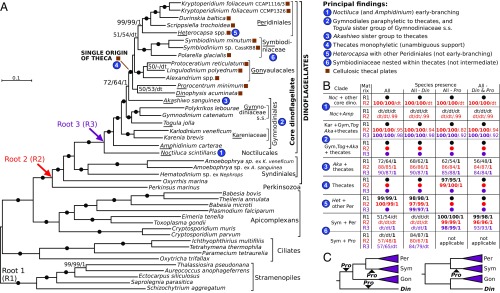
Fig. 1.
Multiprotein phylogeny of dinoflagellates. (A) Best maximum-likelihood tree (IQ-Tree) of dinoflagellates and relatives based on 101-protein dataset (root 1 matrix, 43 species, 29,400 sites). Branches show ultrafast bootstraps (IQ-Tree)/nonparametric bootstraps (RAxML)/posterior probabilities (PhyloBayes) (dash indicates <50/50/0.5 support; filled circles indicate 100/100/1 support; dt indicates a different topology). Roots of alternative matrices (Perkinsus, root 2, 30,780 sites; and Noctiluca, root 3, 30,988 sites) are shown by arrows. (B) Overview of branch supports for principal findings (taxon and matrix abbreviations as underlined in A) in phylogenies of 12 matrices that differ by their root (R1–R3) and species presence (All, All - Din, All - Pro, All - Din & Pro; SI Appendix, Table S1). (C) Two placements of Dinophysis (Din) relative to Gon, Per, and Sym thecates and a variable position of Prorocentrum (Pro) as identified in phylogenies of the 12 matrices (SI Appendix, Table S3, provides tree topology tests).