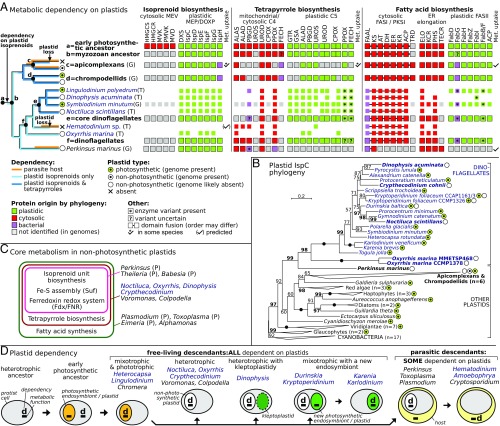
Fig. 3.
Plastid metabolism and dependency in nonphotosyntetic dinoflagellates. (A) Phylogeny-driven reconstruction of plastid and nonplastid variants of core metabolism (isoprenoid, tetrapyrrole, and fatty acid biosynthesis) in genomes (marked as “G”) or transcriptomes (“T”) of dinoflagellates and relatives. Individual enzymes (SI Appendix, Table S5) were classified by protein phylogenies and color-coded as to their presence/absence and origin. The data suggest that Oxyrrhis, Noctiluca, and Dinophysis are metabolically dependent on plastids. Metabolite (Met.) uptake was summarized from the literature. (B) Maximum-likelihood phylogeny (IQ-Tree) reveals IspCs of cyanobacterial origin in nonphotosynthetic dinoflagellates and relatives (bold); ultrafast bootstraps at branches are shown (>50 shown; ≥95 highlighted; filled circles, 100). (C) Three grades in functional organization of core metabolic pathways in nonphotosynthetic plastids in dinoflagellates (blue) and relatives (“P” represents parasites). (D) Model for evolutionary dependency on plastids in dinoflagellates and relatives, which is applicable to other eukaryotes. Ancestral dependency (marked as “d”) on plastid metabolism (loss of cytosolic isoprenoid biosynthesis; later reinforced by the loss of C4 tetrapyrrole biosynthesis in some taxa) led to retention of plastids in all free-living and many parasitic descendants. The dependency can be transferred onto a new plastidial symbiont (Kareniaceae) or host organism (in parasites dependent solely on host-derived metabolites); only the latter leads to an outright loss of the plastid.