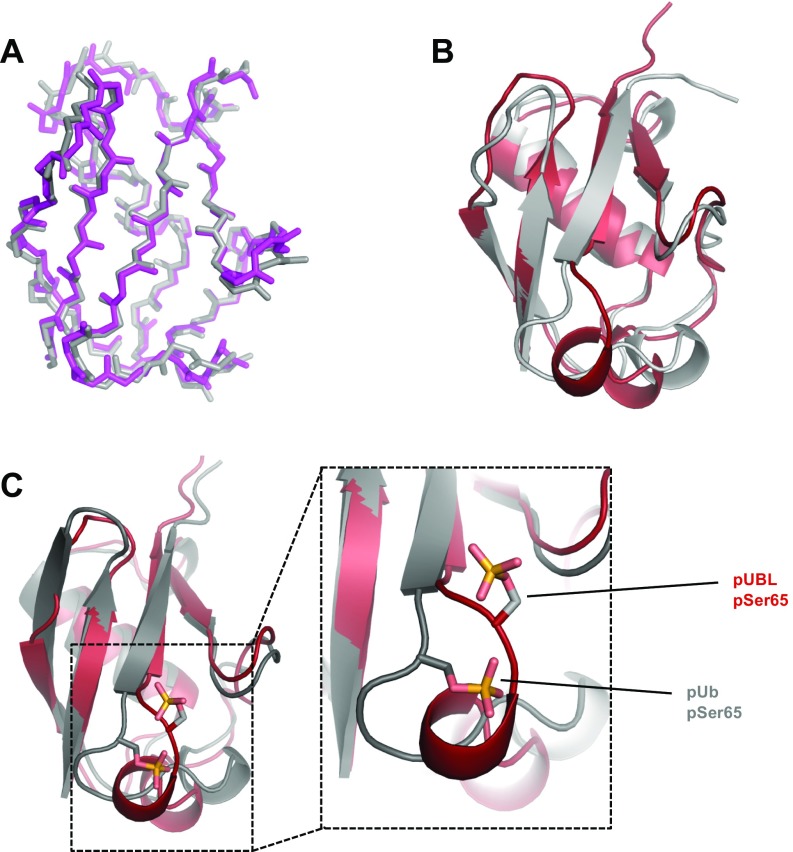
Fig. S2.
Comparisons of existing structures. (A) Backbone atom overlay of existing, unphosphorylated UBL structures. Gray, mouse UBL crystal structure (PDB ID 2ZEQ) (55); magenta, human UBL crystal structure (PDB ID 5C1Z, residues 1–76) (15). Sequence identity between mouse and human orthologues is 89% and backbone RMSD between structures is Å. (B) Superposition of solved pUBL structure (red) and UBL crystal structure (gray, PDB ID 5C1Z). The backbone rmsd between structures is Å. (C) Superposition of solved pUBL structure (red) and pUb crystal structure (gray, PDB ID 4WZP) (21). An expanded view around the phosphorylation site is shown to demonstrate the significant conformational changes observed in UBL vs. Ub upon phosphorylation. pSer65 in ubiquitin maintains a hydrogen to the backbone amide of Q62, whereas pSer65 in UBL is oriented into solvent and facing H68.