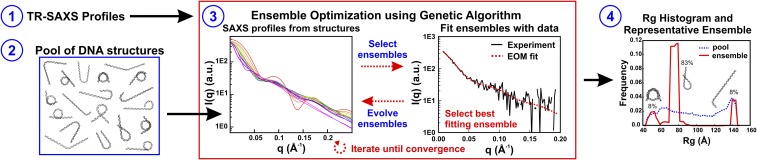
Fig. 3.
Overview of the ensemble optimization method (EOM) used for determining structures. Ensemble optimization (step 3, red box) requires SAXS profiles (step 1) and a pool of DNA structures (step 2) that contains a large number of possible conformations. First, the theoretical SAXS profile for each structure in the pool is calculated using CRYSOL (step 3, Left). A genetic algorithm (GAJOE) randomly selects subsets of these structures, called ensembles, for comparison with the input SAXS data (step 3, Right). Structures from the best-fitting ensembles are propagated into the next generation of ensembles along with some new structures, and this process is repeated (10,000 times) until convergence is achieved. The entire ensemble optimization process is repeated (100 times) to confirm reproducibility and the final ensembles that best represent the data are used to generate histograms of the radius of gyration and to determine the most representative structures for the SAXS profiles (step 4). The example fit and results shown are for the 300-ms time point of NCPs in 1.2 M NaCl under contrast-matched conditions (proteins “invisible” in 50% sucrose).