Fig. 7.
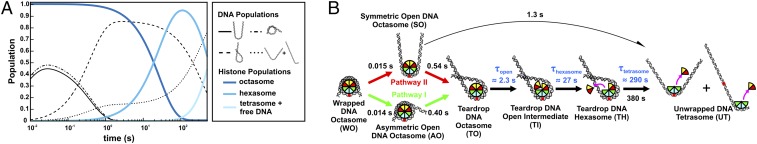
TR-FRET and TR-SAXS analyses reveal hexasome formation at 1.2 M NaCl. (A) Predicted populations of DNA conformational states (black lines) and histone configuration states (blue lines) based on the kinetic rates determined for NCPs at 1.2 M NaCl from the kinetic analysis of EOM models (Figure 6A, see details in Fig. S9) and TR-FRET measurements (Table 1), respectively. (B) NCP disassembly pathway determined from TR-SAXS with histone configurations informed by TR-FRET. Black numbers reflect the SAXS relaxation times (inverse of rates in Fig. S9C). Blue numbers reflect the FRET relaxation times (Table 1). The curved black arrow represents a minor pathway. For simplicity, histone orientations were centered on the dyad when possible.