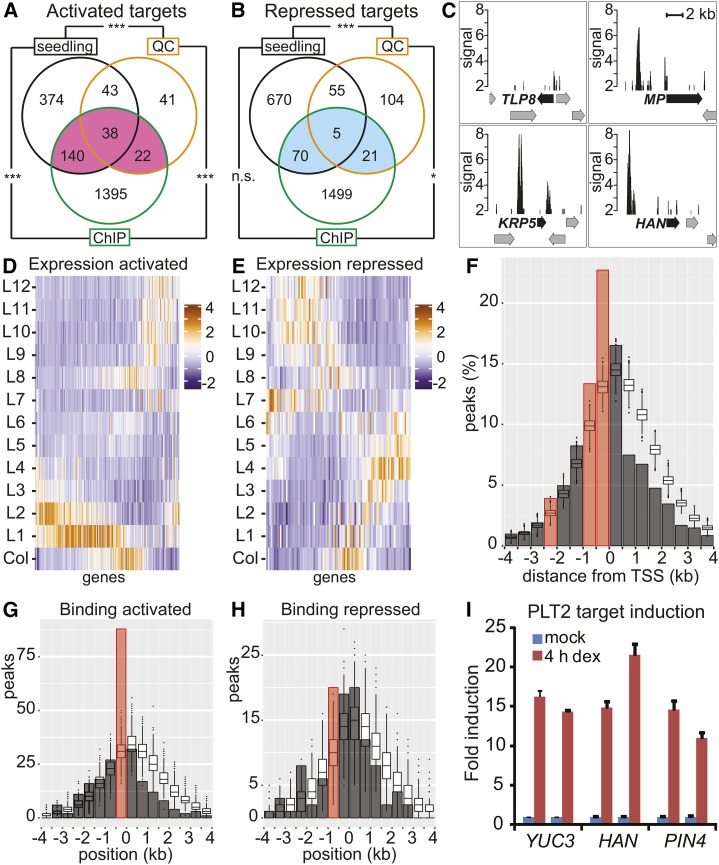
Figure 4.
Genome-Wide Analysis of PLT2 Binding to Target Genes.
(A) and (B) PLT2-activated targets ([A]; pink) and repressed targets ([B]; blue). Overlap between genes bound (ChIP) and transcriptionally regulated by PLT2 (seedlings and QC) represent regulated targets (Supplemental Data Set 5). ***P < 7.6 × 10−29 and *P < 0.005; Fisher’s exact test. n.s., not significant.
(C) PLT2 ChIP-seq profiles of selected regulated targets. Peak signal (CSAR score) plotted with a minimum of 2. Arrows represent genes; black arrows indicate annotated gene for PLT2 ChIP-seq peak.
(D) and (E) Expression profile (z-score) according to the Root Gene Expression Atlas (Brady et al., 2007) of PLT2-activated (D) and -repressed (E) targets from (A) (pink) and (B) (blue), respectively.
(F) Proportion of PLT2 peaks (bars, summit positions were considered) located relative to the closest TSS over the number of peaks assigned within 4 kb (2381 out of a total of 2482 peaks). Box plots show distributions obtained by random selections of genomic positions and assignment to the closest TSS. The x axis displays position of 500-bp genomic bins relative to the TSS within 4 kb. Red bar: significantly enriched over simulated random distribution, FDR < 5%. The shape of the simulated distribution reflects the high gene density of the Arabidopsis genome.
(G) and (H) Number of PLT2 peaks (bars) compared with the simulated distribution of random positions (box plots) relative to the distance from the TSS (0) for PLT2-activated (G) and -repressed (H) targets from (A) (pink) and (B) (blue), respectively. Width of bars and box plots represent 500-bp genomic bins around the TSS. Red bar indicates significantly enriched over simulated random distribution, FDR < 5%.
(I) qRT-PCR analysis of PLT2 target induction. Expression levels of YUC3, HAN, and PIN4 transcripts in root tips after 4 h DEX induction of Pro35S:PLT2-GR compared with the mock-treated condition. Data represent biological duplicates with each time point consisting of technical triplicates. Error bars indicate se.