Figure 1.
PG-M48 Is Localized in PGs and Has Zn-Metallo-Dependent Endopeptidase Activity Cleaving Upstream of Hydrophobic Residues.
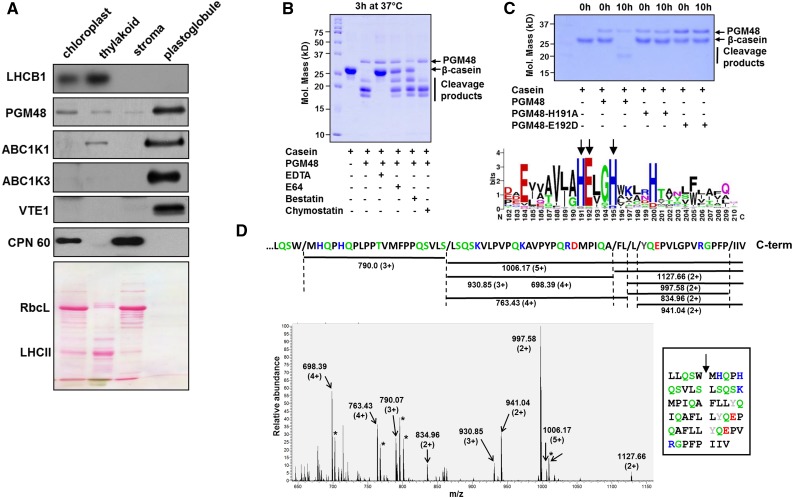
(A) Proteomes from isolated chloroplasts, thylakoid membranes, stroma, and PGs were separated by SDS-PAGE, transferred to membranes, and blotted with specific antisera against PG core proteins PGM48, ABC1K1, ABC1K3, and VTE1, as well as LHCB1 (marker for thylakoid membrane) and CPN60 (marker for stroma). Protein (15 µg) was loaded for chloroplast, stroma, and thylakoid fractions and ∼5 µg protein was loaded for PGs; due to the extremely high lipid/protein content, protein concentrations are difficult to measure accurately. The Ponceau-stained membrane (lower panel) serves as loaded control. RBCL, Rubisco large subunit (also marker for stromal proteome); LHCII, family of major LHCII proteins.
(B) Recombinant PGM48 (1 µg) was incubated for 3 h at 37°C with β-casein (4 µg) to determine the peptidase activity and the effects of peptidase inhibitors E64 (1 mM), bestatin (1 mM), chymostatin (1 mM), and the metal chelator EDTA (5 mM). EDTA inhibited PGM48 peptidase activity completely.
(C) Metal dependency of PGM48. Upper panel shows in vitro proteolytic activity of recombinant PGM48, PGM48-H191A, and PGM48-E192D mutated in their predicted metal binding site (HExxH). PGM48, but not mutated PGM48, shows peptidase activity. The lower panel shows a sequence logo for the conserved metal binding motif of 11 PGM48 homologs in angiosperms (for a complete list of homologs, see Supplemental Table 1). Color coding: green: Thr, Gly, Ser, Tyr, and Cys, uncharged, polar; pink: Asn and Gln, uncharged polar amine; red: Asp and Glu, acidic; blue: Lys, Arg, and His, basic; black: Ala, Val, Leu, Ile, Pro, Phe, Trp, and Met, apolar.
(D) PGM48 cleavage sites of β-casein as determined by mass spectrometry. The lower panel shows a high-resolution MS spectrum (from the Orbitrap portion of the LTQ-Orbitrap) of a casein digest by PGM48. Positively charged peptides matching to casein peptide fragments are marked; m/z values and charge states are indicated. The upper portion shows the C-terminal portion of β-casein and the identified peptides as indicated by horizontal lines with the respective m/z value and associated charge states. The inset shows the inferred cleavage sites with the β-casein sequence. Collectively, this shows that PGM48 prefers cleavage upstream of hydrophobic residues: methionine (M), phenylalanine (F), leucine (L), isoleucine (I) (in black), or tyrosine (Y) (in gray). Other colors: green: Q,S – uncharged, polar; D,E – acidic in red; K,R,H – basic in blue; remaining residues in black.