Figure 2.
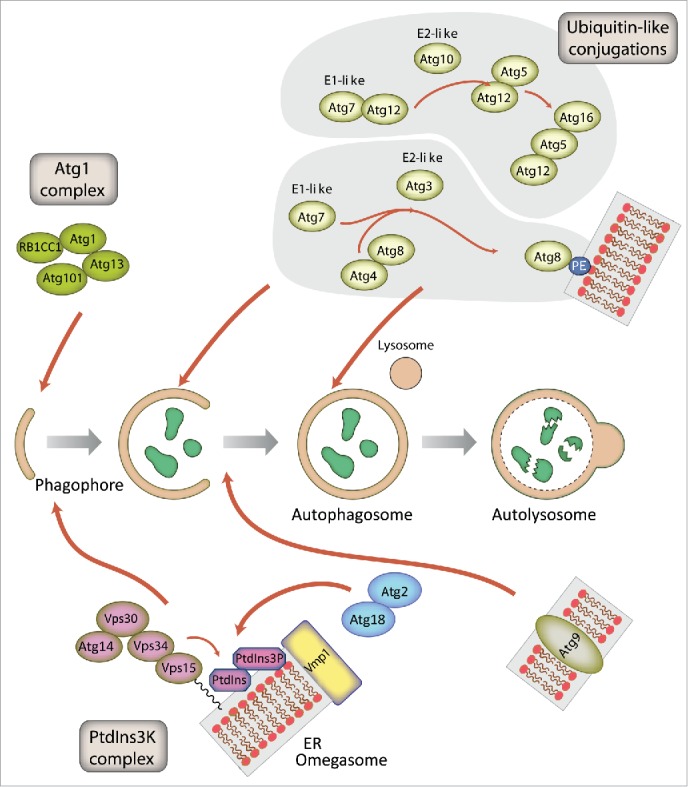
Schematic representation of autophagosome formation and the proteins involved in D. discoideum. The autophagic proteins were identified by sequence homology with those described in yeast and mammalian cells. The predicted complexes have been organized using the information available from the yeast S. cerevisiae, mammalian cells and D. discoideum. The inductive stage depends on the serine/threonine Atg1 kinase complex and the class III PtdIns3K complex. The latter complex generates the signaling lipid PtdIns3P, which is essential for the recruitment of Atg18 and Atg2. The elongation of the phagophore membrane requires 2 ubiquitin-like (Ubl) conjugation systems (upper right). In the first conjugation reaction Atg12 is covalently bound to Atg5. Atg12–Atg5 interacts with Atg16 and localizes to the phagophore membrane, from which it regulates the second conjugation reaction that attaches the protein Atg8 (known as LC3 in mammals) to phosphatidylethanolamine (PE) of the expanding phagophore membrane. Atg8 initially localizes to both sides of the phagophore and remains inside the completed autophagosome until fusion with the lysosome occurs; Atg8 on the cytosolic side of the autophagosome is cleaved from PE by Atg4 and recycled, whereas Atg8–PE of the lumenal leaflet of the bilayer is degraded. Atg9-containing vesicles supply membrane for phagophore expansion. Recent advances have clarified the composition of the Atg1 complex and the role of Vmp1 as a key protein in the origin of the autophagosomes from the ER.
