Figure 5.
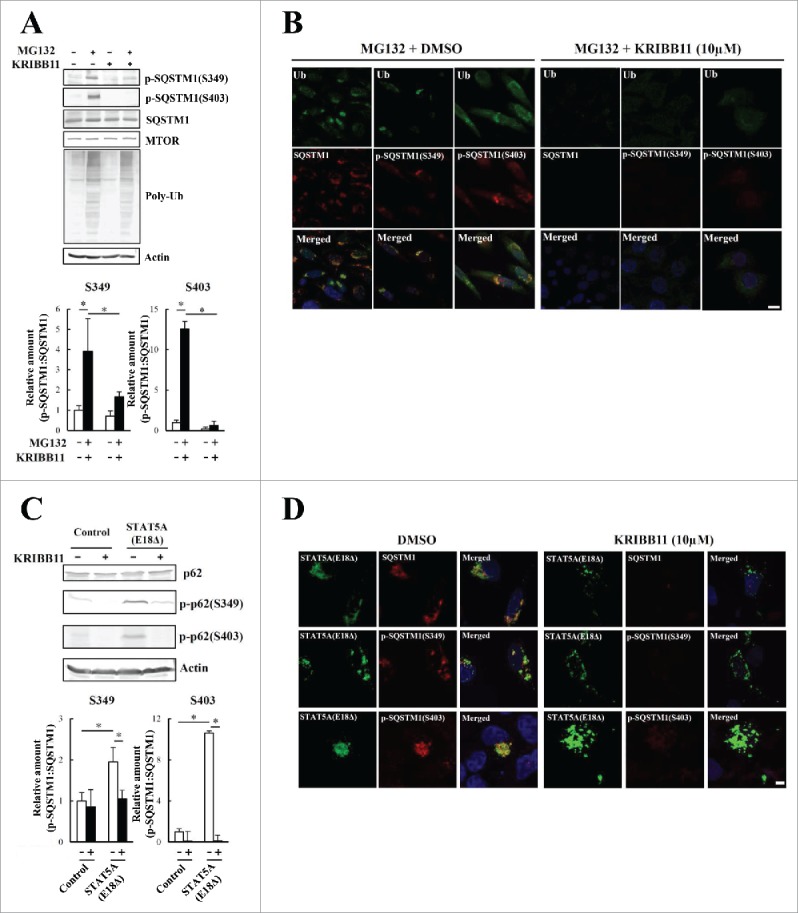
Phosphorylation of S349 and S403 on SQSTM1 is suppressed by a KRIBB11 HSF1 inhibitor. (A) HeLa cells were treated with 10 µM MG132 and 50 µM KRIBB11 for 12 h. Cell lysates were subjected to immunoblot analysis. Band intensities were measured, and phosphorylated SQSTM1 values were normalized to total SQSTM1. The data are reported as means ± SD (n = 4). Statistical analyses were performed using one-way ANOVA, followed by the Tukey post-hoc test. *P < 0.01. (B) Colocalization of SQSTM1 with ubiquitinated inclusions were immunocytochemically assessed in cells treated with MG132 alone (left) or MG132 and KRIBB11 (right). Cell nuclei were counterstained blue with DAPI. Scale bar: 10 μm. (C) HeLa cells were transiently transfected with mock control or pEGFP-STAT5A(E18Δ) (STAT5A[E18Δ]), and then treated (+) with or without (−) 50 µM KRIBB11. After 12 h, cell lysates were examined by immunoblot analysis. Band intensities of S349- and S403-phosphorylated SQSTM1 were measured, and the data are reported as means ± SD (n = 4). Statistical analyses were performed using one-way ANOVA, followed by the Tukey post-hoc test. *P < 0.01, **P < 0.05. (D) Colocalization of SQSTM1 with EGFP-STAT5A(E18Δ) were immunocytochemically assessed in cells treated with DMSO control (left) or KRIBB11 (right). Cell nuclei were counterstained blue with DAPI. Scale bar: 10 μm.
