Figure 3. Step-by-Step Reconstruction and Structure of Double-Stranded Fibrin Oligomer FO2/3.
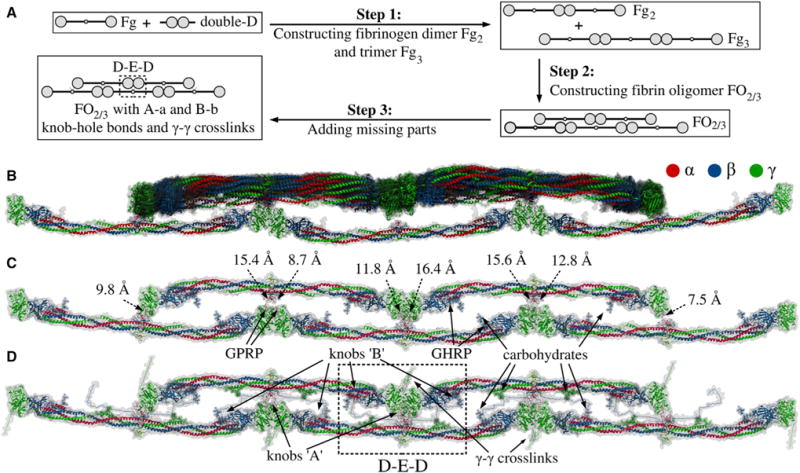
(A) Steps 1–3 summarize the procedure used to reconstruct double-stranded fibrin oligomer FO2/3 (see Experimental Procedures). As the input, we use the atomic structural models of human fibrinogen (PDB: 3GHG; Kollman et al., 2009) and double-D fragment (PDB: 1N86; Yang et al., 2002). These structures are utilized in step 1 to create fibrinogen dimer Fg2 and trimer Fg3, which are used in step 2 to construct a double-stranded fibrin oligomer FO2/3 with two/three monomers in the upper/lower strand. The initial model of FO2/3 does not contain knobs ‘A’ and ‘B’ and the γ-γ crosslinks, which are incorporated in step 3. To remove steric clashes and to minimize the potential energy of the obtained conformation of FO2/3, this step is followed by energy minimization. See also Figure S2 for geometric constraints used in Monte Carlo docking.
(B) Superposition of all 14 successfully reconstructed structures from the Monte Carlo docking (see Supplemental Information).
(C and D) Representative structures of FO2/3 before and after the incorporation of missing residues: knobs ‘A’ and ‘B’ and g-g crosslinks (see Data S2). (C) also shows the distances for all eight constraints due to A-a knob-hole bonds. See also Figure S2 and Data S2.
