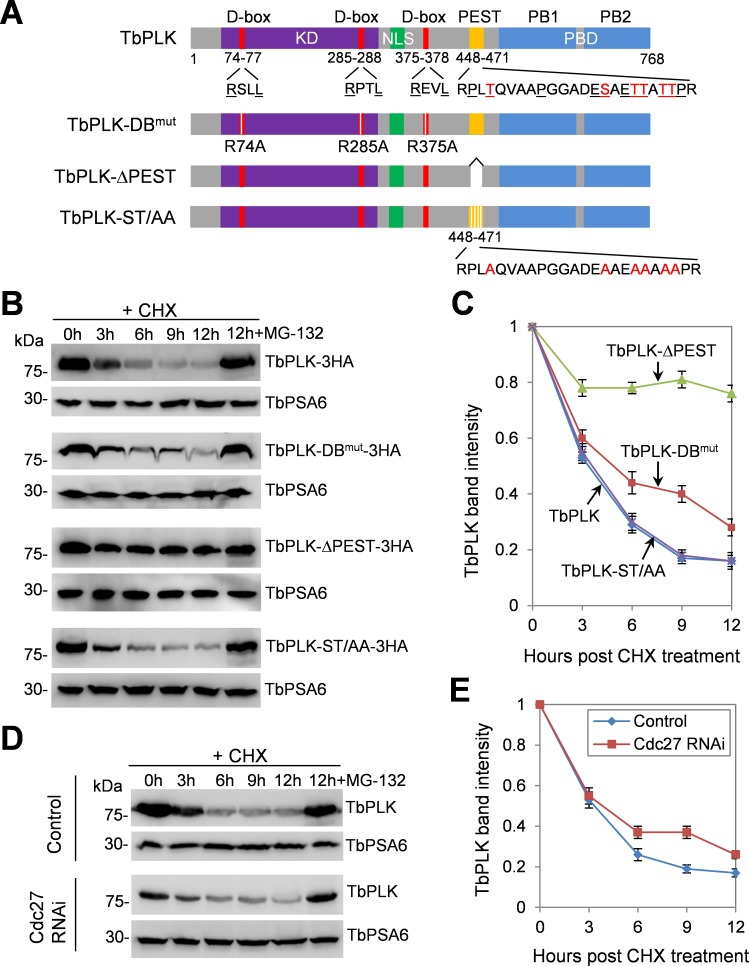
Fig 1. Degradation of TbPLK is mediated by destruction box and PEST motif.
(A). Schematic drawing of the domains in TbPLK and the various mutants of TbPLK expressed for protein stability assay. DB, destruction box or D-box; KD, kinase domain; NLS, nuclear localization signal sequence; PEST, proline (P), glutamic acid (E), serine (S) and threonine (T)-enriched sequence; PBD, Polo-box domain; PB1 and PB2, Polo-boxes 1 and 2. The signature arginine (R) and leucine (L) residues in the three D-boxes, and the proline (P), glutamate (E), serine (S) and threonine (T) residues in the PEST motif are underlined. The phosphorylated serine and threonine residues in the PEST motif are highlighted in red. (B). Degradation of ectopically overexpressed TbPLK and its D-box mutant and PEST-deletion mutant. Cells overexpressing 3HA-tagged TbPLK or each of the TbPLK mutants were treated with cycloheximide, and time-course samples (equal numbers of cells) were collected for Western blotting with anti-HA antibody. In a separate cell sample, MG-132 was added together with cycloheximide and incubated for 12 h (12h+MG-132). TbPSA6, the T. brucei proteasome subunit alpha-6, served as the loading control. (C). Quantification of TbPLK band intensity from panel B. TbPLK band intensity was measured with ImageJ, and normalized with the band intensity of TbPSA6. Error bars represent S.D. calculated from three independent experiments. (D). Degradation of TbPLK in control and Cdc27 RNAi cells, which was induced for 72 h. Cells were treated with cycloheximide for up to 12 h, and time-course samples were collected for Western blotting with anti-TbPLK antibody. The level of TbPSA6 served as the loading control. (E). Quantification of TbPLK band intensity from panel D. TbPLK band intensity was measured with ImageJ, and normalized with the band intensity of TbPSA6. Error bars indicate S.D. calculated from three independent experiments.