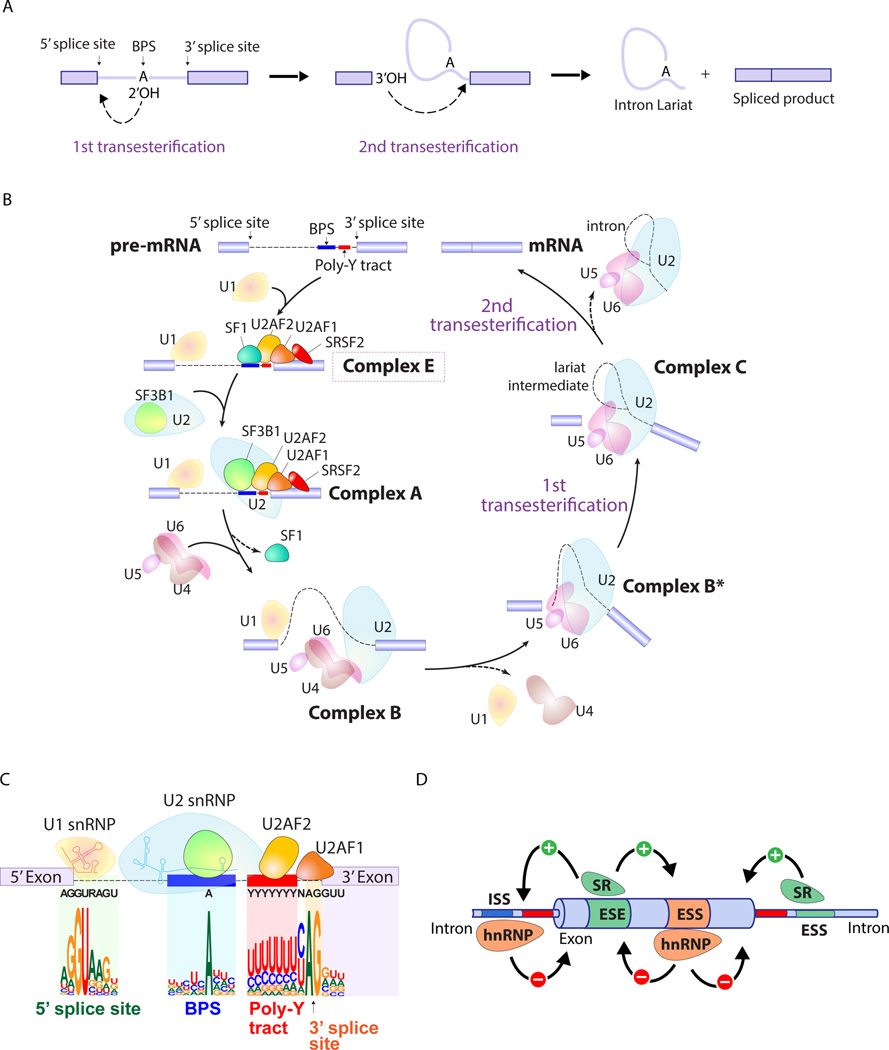
Figure 1. Splicing catalysis, the spliceosome assembly pathway, and mechanisms of splice site selection.
(A) Diagram of the 2 sequential transesterification reactions that represent the crucial catalytic steps in intron removal during splicing. An adenine nucleotide (termed the "invariant adenine") of the branch point sequence (BPS) initiates the first transesterification and generates a free 5' exon and an intron-3' exon lariat. The 3' end hydroxyl of the free 5' exon then attacks the intron-3' exon junction, completing the splice and releasing a lariat RNA intron. (B) Pre-mRNA splicing is a dynamic process that involves several distinct spliceosomal complexes. The earliest complex (complex E) is established by binding of (i) U1 snRNP to the the 5’ splice site (SS), (ii) splicing factor 1 (SF1) to the BPS, (iii) U2AF2 (also known as U2AF65) to the polypyrimidine tract, (iv) U2AF1 (also known as U2AF35) to the 3’ SS. Formation of complex E in turn enhances the recruitment of U2 snRNP to the BPS and leads to the formation of complex A. SF3B1, a component of U2 snRNP, is involved in the binding to the BPS. The pre-assembled U4/U6.U5 tri-snRNP complex joins and the U1/U4 snRNPs are released to form the catalytically active complex B (complex B*), followed by the further conformational rearrangements that results in the formation of complex C. Complexes B and C catalyze the first and second esterification reactions, respectively, and mediate excision of the intron and ligation of the proximal and distal exon to synthesize mature mRNA. (C) A focus on complex E highlights consensus sequence elements recognized by U1 and U2 snRNPs as well as the U2AF complex. An intron is defined via (i) the 5’ SS, (ii) the 3’ SS, (iii) the branch point sequence (BPS), and (iv) the polypyrimidine (Poly-Y) tract. The definition of an intron depends on recognition of the 5’ SS and BPS by U1 and U2 snRNPs, respectively. The consensus sequences shown are those recognized by the major (U2-dependent spliceosome) which processes >95% of introns (as opposed to the minor U12-dependent spliceosome which recognizes different consensus sequences than those shown here). (D) In addition to sequences in mRNA recognized by the core spliceosome and the U2AF complex, accessory splicing regulatory proteins are essential in promoting or repressing splice site usage. Members of the serine/arginine (SR) family proteins control the pattern of alternative splicing by recognizing specific sequences in pre-mRNA named exonic and intronic splicing enhancers (ESE and ISE). SR proteins generally act as enhancers of splicing from nearby splice sites by interacting ESE and ISE, while heterozygous nuclear ribonucleoprotein particle (hnRNP) suppresses splicing by interacting with exonic and intronic splicing silencers (ESS and ISS).