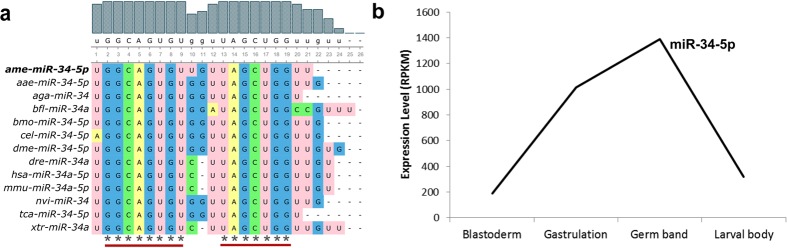
Figure 3. miR-34 emerges as a putative regulator of honey bee embryogenesis.
(a) Multiple alignment of miR-34-5p in animal species. Gray bars indicate conservation degree for each base in the miR-34-5p. The nucleotide sequence underneath the bars represents the consensus sequence (the upper case letters correspond to the bases conserved in all analyzed species. The red bars indicate the conserved seed region (2–9 nt). (b) Transcripts levels of miR-34-5p in honey bee embryogenesis assessed by RNA-seq. aee: Aedes aegypti; aga: Anopheles gambiae; ame: A. mellifera; bfl: Branchiostoma floridae; bmo: Bombyx mori; cel: Caenorhabditis elegans; dme: D. melanogaster; dre: Danio rerio; hsa: Homo sapiens; mmu: Mus musculus; nvi: Nasonia vitripennis; tca: Tribolium castaneum; xtr: Xenopus tropicalis.