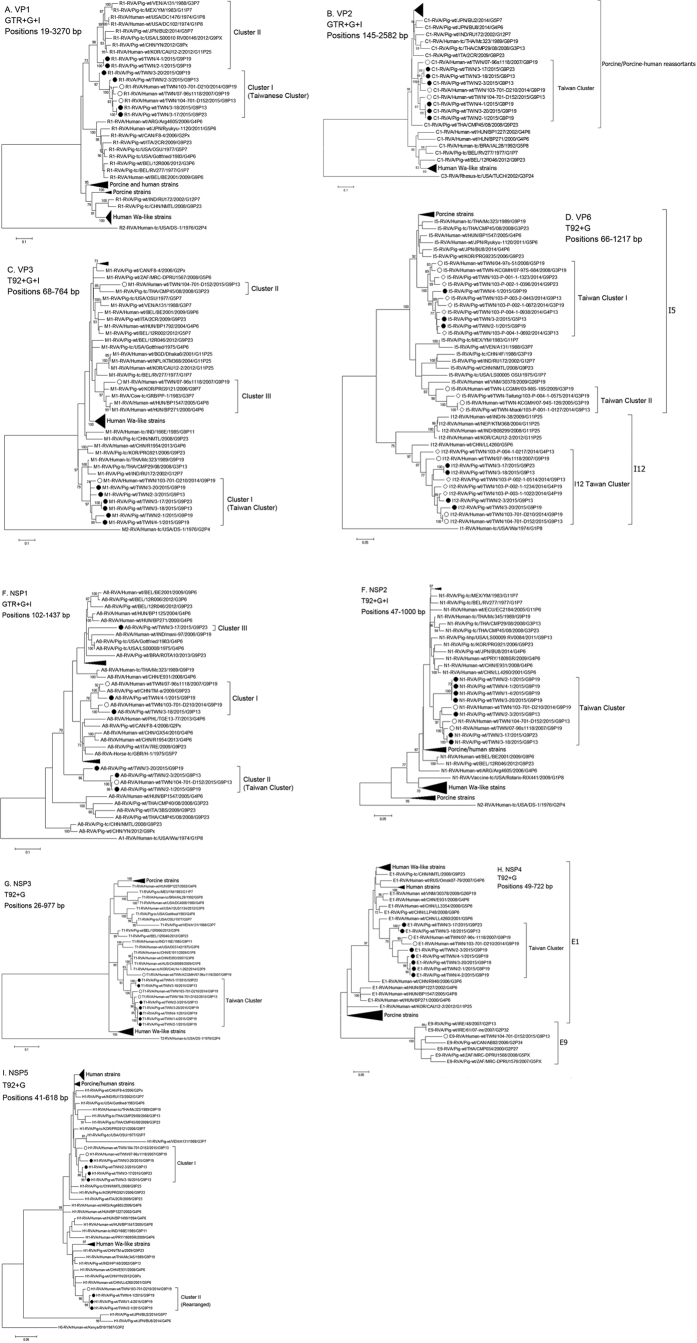
Figure 5.
Condensed phylogenetic trees of (A) VP1, (B) VP2, (C) VP3, (D) VP6, (E) NSP1, (F) NSP2, (G) NSP3, (H) NSP4, and (I) NSP5 were constructed with MEGA6 using neighbor-joining (maximum composite likelihood) or maximum likelihood method with 1,000 bootstrap replicates using the best-fit substitution models determined by the built-in model test application: GTR = General Time Reversible, T92 = Tamura 92, HKY = Hasegawa-Kishino-Yano 85, TN93 = Tamura-Nei 93, G = gamme sites, I = invariant sites. The region within each of the gene segment used for each analysis is also shown. Open circles indicate Taiwanese human strains, closed circles denote Taiwanese porcine strains from 2015, open diamonds denote Taiwanese porcine strains from 2014.