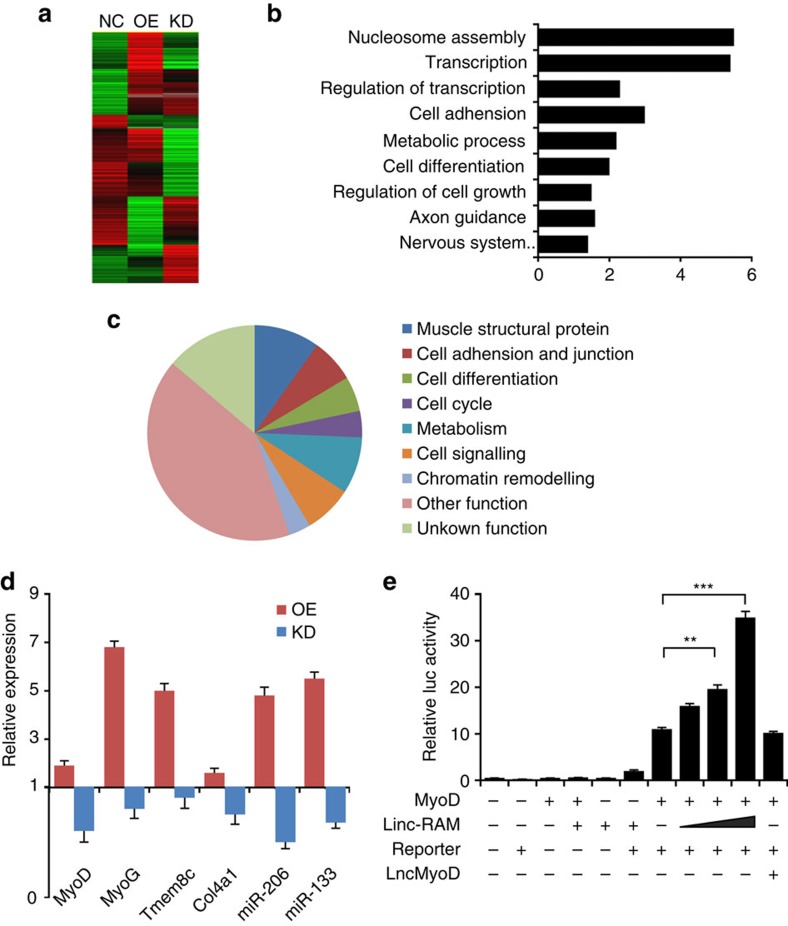
Figure 4. Linc-RAM acts as a regulatory lncRNA enhancer of MyoD in regulating expression of myogenic genes.
(a) RNA sequencing analyses were applied to stable Linc-RAM-overexpressing (OE) and Linc-RAM knockdown (KD) C2C12 cell lines; cells stably infected with vector alone (NC) served as controls. The heatmap shows hierarchical clusters for 852 differentially expressed genes, which were designated Linc–RAM-affected/targeted genes. Red, upregulated; green downregulated (cutoff, ≥1.5-fold change; P≤0.005). (b) Enriched GO terms for Linc–RAM-affected genes. The y axis shows GO terms and the x axis shows statistical significance (negative logarithm of P value). (c) MyoD ChIP-seq data were applied to analyse whether Linc-RAM-affected genes were also regulated by MyoD. The pie chart demonstrates GO term classifications for 151 Linc-RAM-affected genes with at least one MyoD-binding peak in the promoter region. (d) Differentially expressed genes were validated by RT–qPCR. (e) Luciferase reporter gene assay to measure MyoG promoter activity in fibroblasts cotransfected with MyoD and different amounts of Linc-RAM. LncMyoD, a non-related long non-coding RNA, served as negative control. The data are presented as mean±s.e.m. from three independent experiments. The statistical significance was calculated with the t-test, **P<0.01, ***P<0.001.