Figure 4.
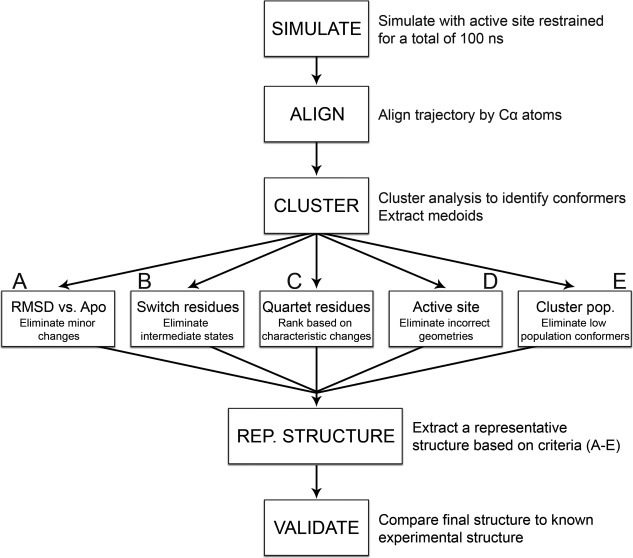
Flowchart summarizing the methodology. Initial apo structure is phosphorylated and simulated with active site restraints. The resulting combined ensemble is aligned to remove translational and rotational fluctuation. Conformers are then clustered together based on similarity using k‐medoids based algorithm. Candidate structures (medoids) are extracted for each cluster. Each medoid is analyzed to identify the candidate most likely to resemble a phosphorylated crystal structure using the following criteria: (A) Candidates are aligned to their apo structure to eliminate medoids showing little to no conformational changes (∼3.5 Å cutoff). B: Switch residue positions are analyzed. Medoids lacking fully buried aromatic side chains and hydrogen‐bonded Thr/Ser residues (2.9 Å cutoff) are ignored. C: Quartet residue positions are measured and compared between medoids. D: Active site geometry is checked. Medoids lacking fully conserved active site arrangements are ignored. E: Medoids from transient clusters are eliminated (∼1500 conformer cutoff). Using these criteria, a final representative structure is identified. This structure is then validated against existing structural data.
