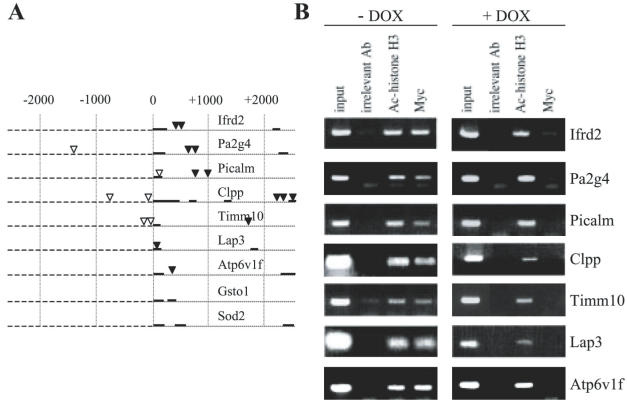
Figure 5.
Myc binds to the consensus CACGTG sequences in target genes. (A) Mouse genomic DNA sequences for indicated genes, obtained from Celera Discovery System, were searched for CACGTG motifs located within a distance of 2 kb on either side of transcription start site. Triangles represent consensus binding sites, whereas full triangles symbolize consensus CACGTG sequences further analyzed by ChIP. Full lines represent exons, dark dashed lines introns and light dashed lines non-transcribed genomic sequence. (B) Cells treated or not treated with doxycycline, were cross-linked with formaldehyde and the chromatin was immunoprecipitated using anti-Myc or anti-AC histone H3 antibody. After reverse cross-linking, co-immunoprecipitated DNA was analyzed by PCR using primers specific for the regions containing CACGTG motifs into indicated Myc target genes. Lyn or PLCγ antibodies were used as a negative control (irrelevant antibody). Representative data obtained with the cell line 5522 B are shown.