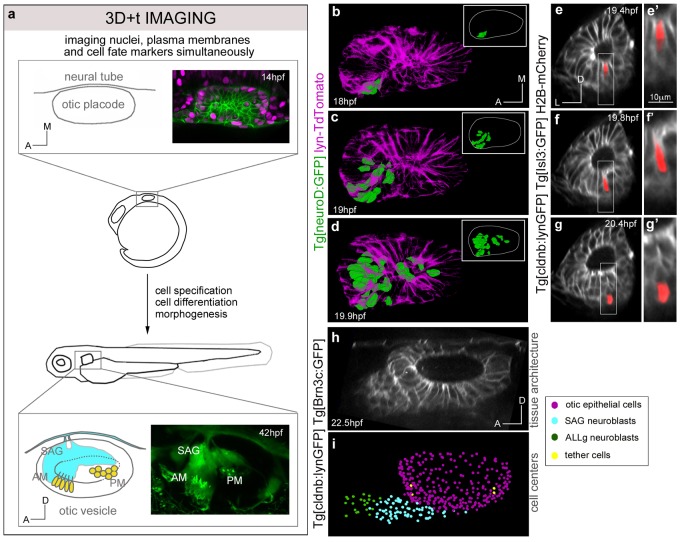
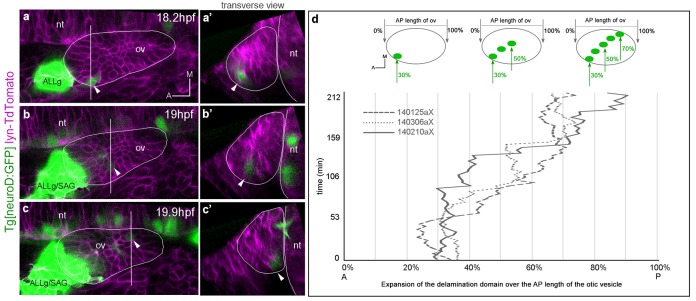
Figure 1. Expansion of the neuroblast delamination domain and formation of the SAG rudiment.
(a) Overview of the imaging and image processing strategy: inner ears of zebrafish embryos stained for cell membrane, nucleus and cell fate markers were imaged between 14-42 hpf. Image datasets were processed by nucleus center detection, cell tracking and cell shape segmentation. Data were validated and curated (Figure 1—figure supplement 1). (b–d) Time-lapse stills showing the posterior expansion of the neuroblast delamination domain over time; 3D-rendering of segmented epithelial neuroblasts (green) in context of the otic structure (plasma membranes in magenta) at indicated times; insets display only the segmented delamination domain with the otic vesicle contour in white. ID Dataset: 140210aX; see Figure 1—figure supplement 2d for additional analyses. (e–g) Time-lapse stills showing a segmented delaminating neuroblast (red; Video 2); (e’–g’) magnifications of framed regions in (e–g). ID Dataset: 140426aX. (h–i) Still images from Video 1 displaying: otic tissue architecture (h), and cellular distribution (i) upon SAG formation. Reconstructed cell centers are color-coded according to cell position/identity (see legend). ID Dataset: 140423aX. SAG/ALLg, statoacoustic/anterior lateral line ganglia. AM/PM, anterior/posterior maculae.
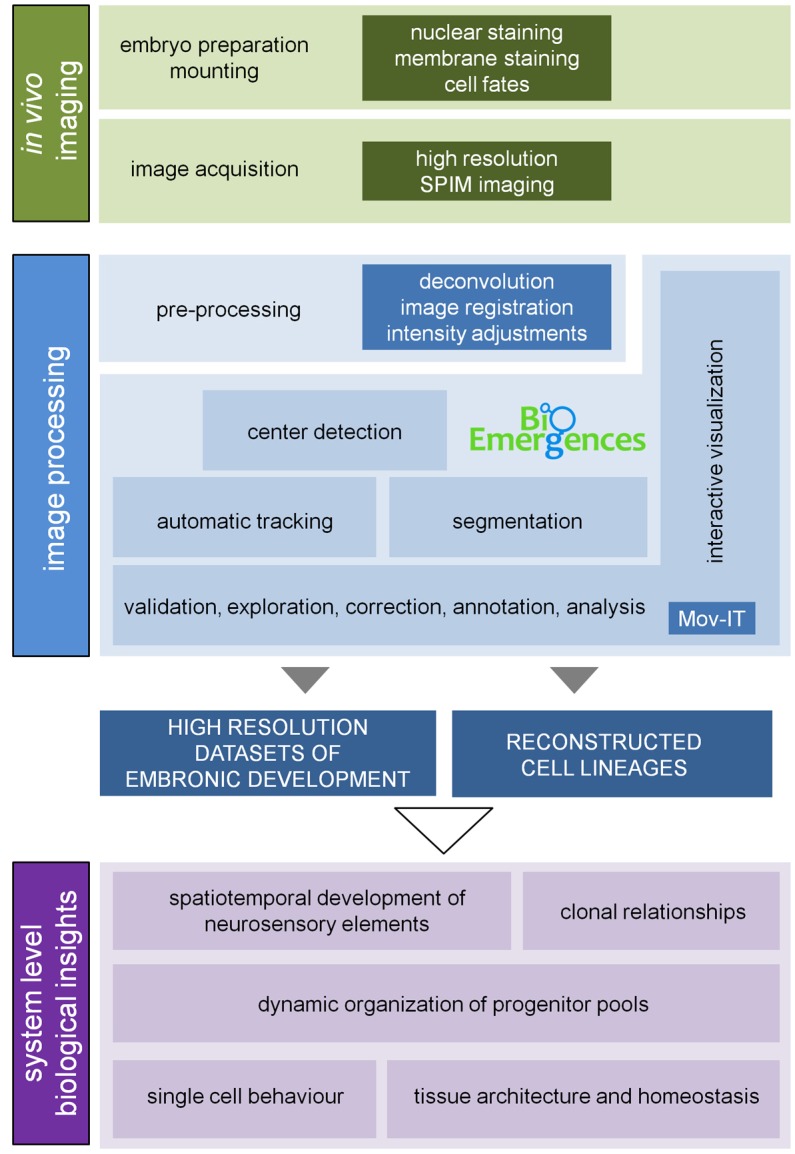
Figure 1—figure supplement 1. 3D+time image analysis pipeline.