Figure 4. CP1 promotes the decay of CRU proteins.
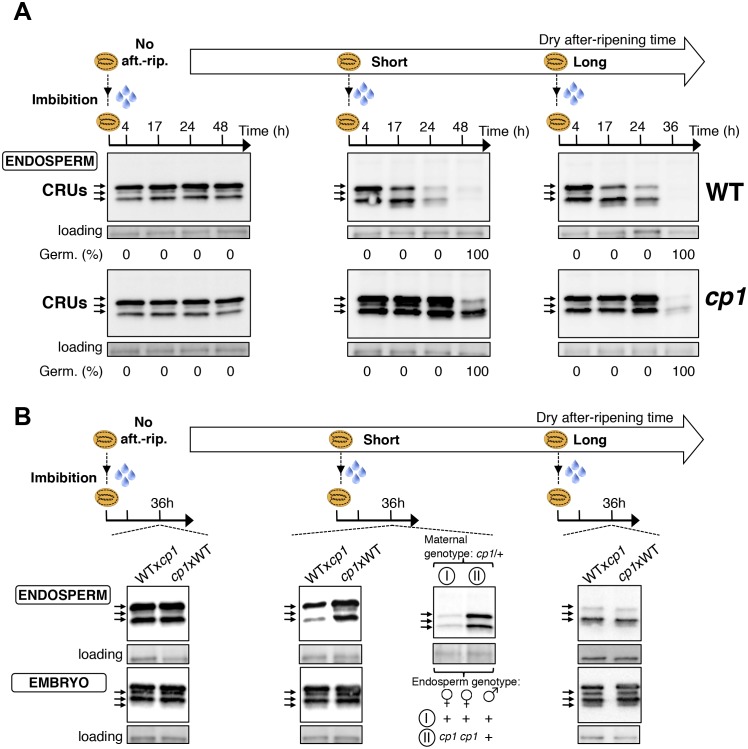
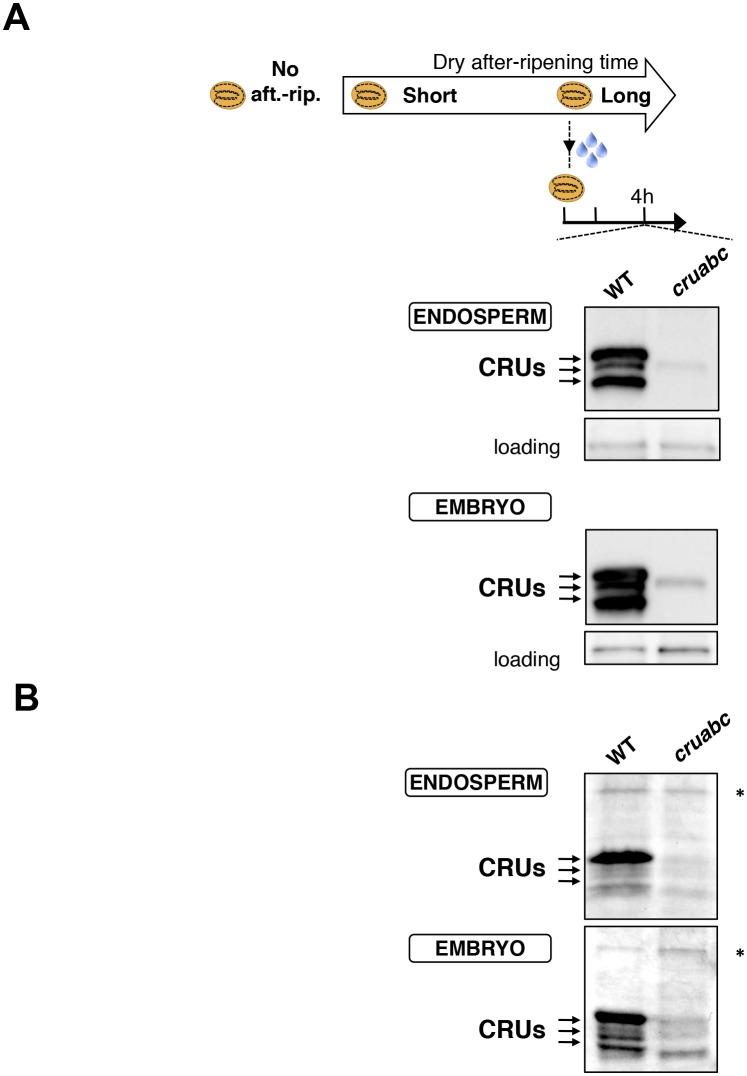
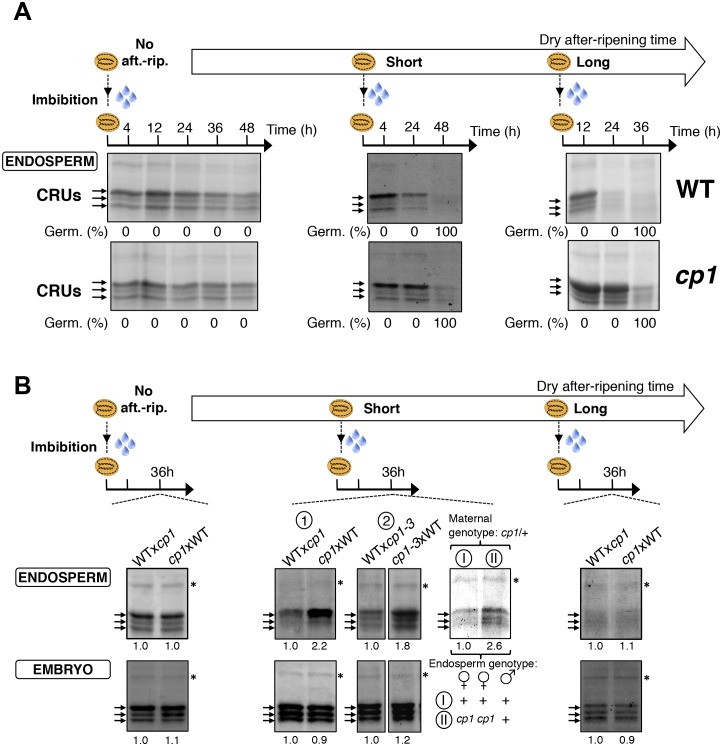
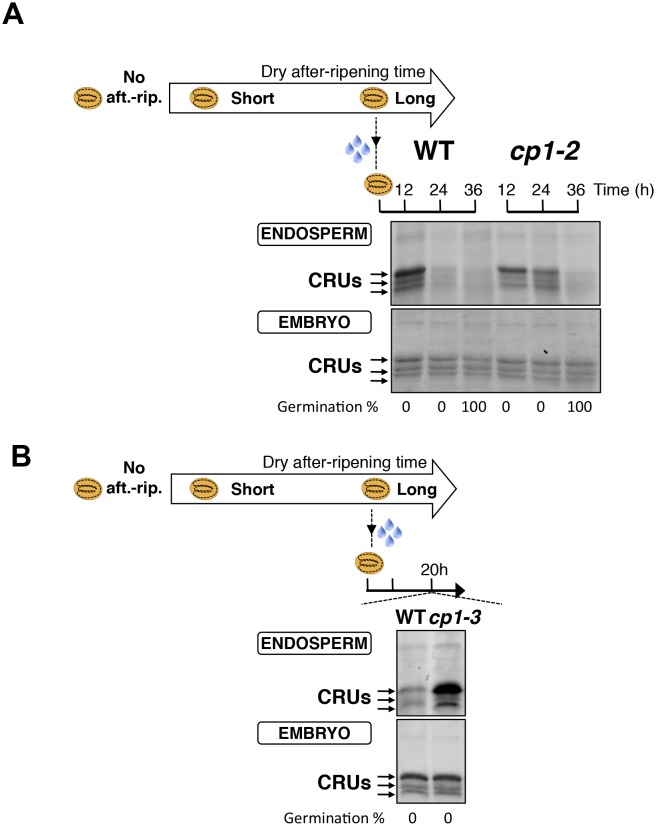
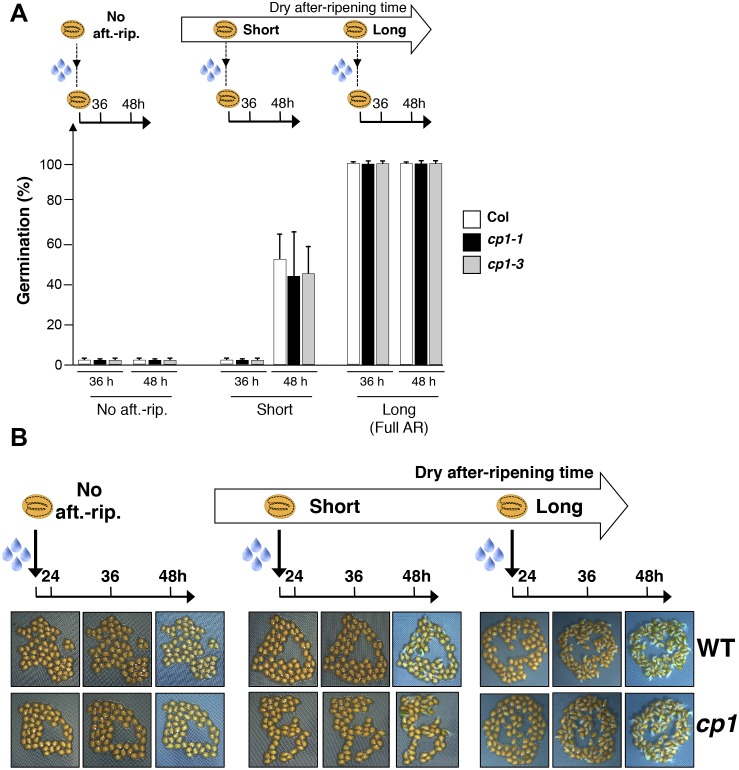
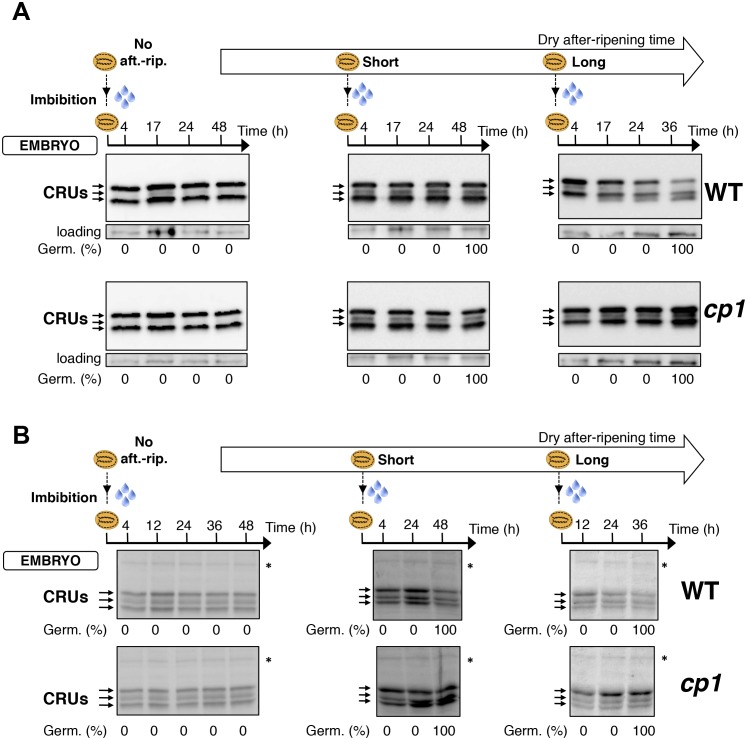
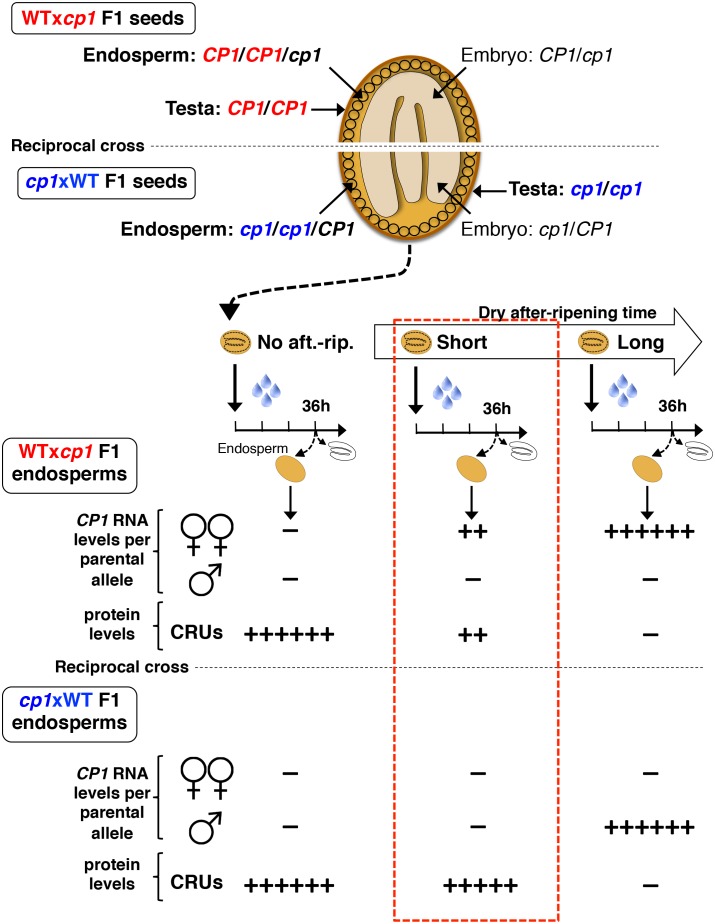
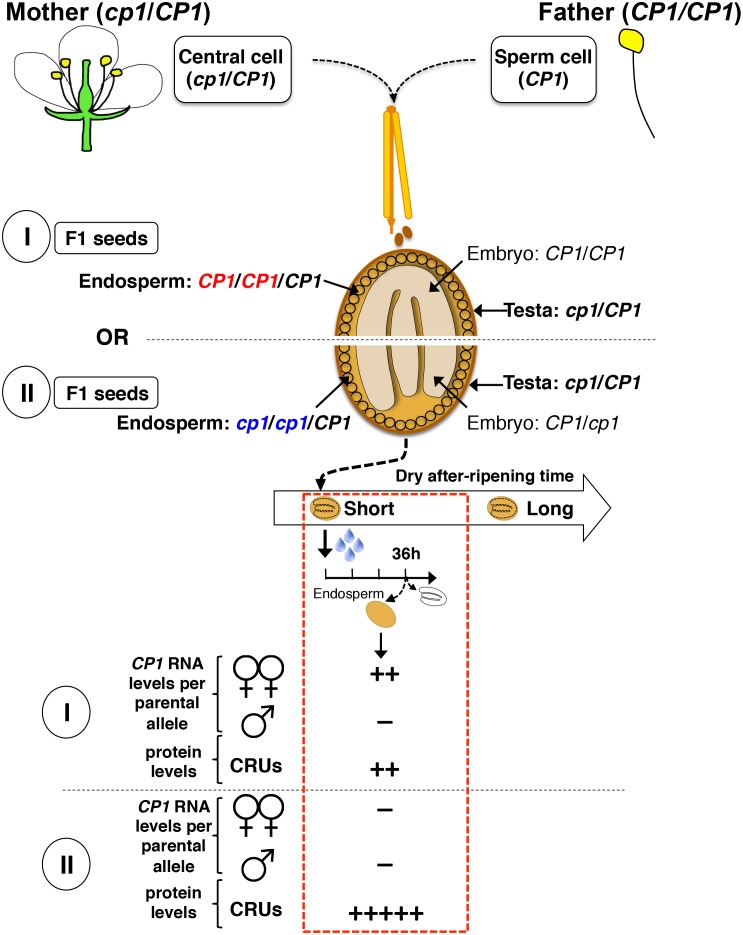
CRU protein decay is controlled through CP1 maternal gametophytic alleles. (A) Endosperms from non-after-ripened (No aft.-rip.) WT and cp1 seeds or from WT and cp1 seeds after-ripened for short (three days) or long (two months) time periods were dissected at the indicated times after seed imbibition. Proteins isolated from the same number of WT (Col) or cp1 endosperms were separated on SDS PAGE gels and probed with 12S antibody serum. The specificity of the antibody was confirmed using protein extracts isolated from the endosperm and embryo of the cruabc mutant (Figure 4—figure supplement 1). For a given after-ripening time, WT and cp1 endosperm proteins were separated on the same SDS PAGE gel. An aspecific band is used as a loading control. Percent germination at each time-point after seed imbibition is indicated. Arrows indicate highly abundant cruciferin (CRU) proteins. (B) WTxcp1 F1 and cp1xWT F1 seeds (obtained by reciprocally crossing WT (Col) and cp1 plants) were subject to dry after-ripening treatments as in (A). For each after-ripening treatment, endosperms and embryos were dissected 36 hr after seed imbibition. Proteins isolated from the same number of endosperms or embryos were processed as in (A). Seeds obtained after crossing cp1/+ heterozygous mother plants with Col WT (+/+) pollen were after-ripened for a short time period (three days). Endosperms were dissected 36 hr after imbibition, whereas embryos were further cultured for later genotyping in order to distinguish endosperms according to their (i) +/+/+ or (ii) cp1/cp1/+ genotype. Endosperms with the same genotype were pooled. Proteins extracted from the two endosperm pools were processed as in (A).