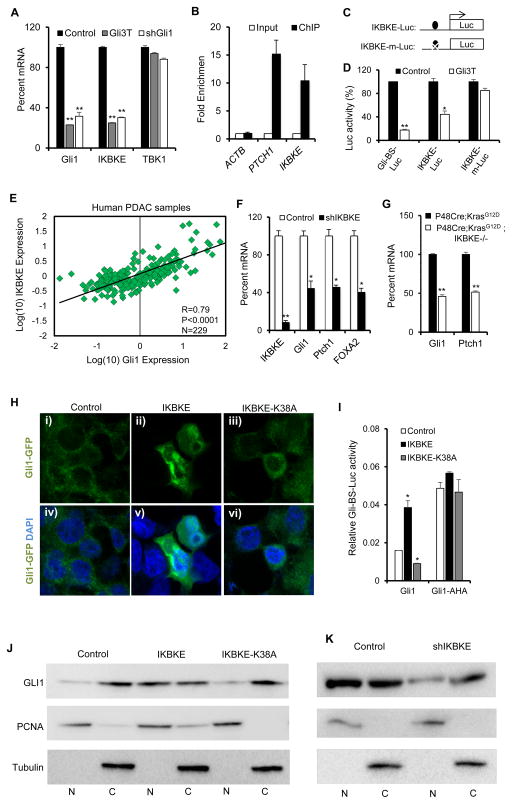
Figure 2. Reciprocal IKBKE-Gli1 interaction in PDAC cells.
(A) qPCR analysis in Panc-1 cells showing GLI1, IKBKE, and TBK1 mRNA expression after inhibition of GLI activity using a dominant negative GLI (Gli3T), or shRNA targeting GLI1. (B) Quantitative PCR of DNA enriched using ChIP against Flag-tag in Panc-1 cells infected with Gli3T-Flag indicates significant enrichment of IKBKE promoter region as well as the promoter of a known GLI target gene (PTCH1), but not control ACTB promoter region. (C) Schematic showing location of GLI binding site and mutated GLI binding site upstream of luciferase reporters. (D) Relative luciferase activity in Panc-1 cells expressing Gli-BS luciferase reporter (Gli-BS-Luc), IKBKE luciferase containing the wild type GLI binding site (IKBKE-Luc), and IKBKE luciferase carrying a mutated GLI binding site (IKBKE-m-Luc), with or without GLi3T ectopic expression. (E) Correlation of GLI1 and IKBKE mRNA expression in human PDAC tissue samples (n=229). Pearson coefficient of R = 0.79 indicates high degree of correlation between GLI1, and IKBKE expression in the tumors. (F) Quantitative RT-PCR analysis of transcription of IKBKE and the known GLI target genes, GLI1, PTCH1, and FOXA2, in Panc-1 cells with IKBKE knockdown. (G) Quantitative RT-PCR in tissue samples indicates significantly reduced expression of GLI target genes, Gli1 and Ptch1, in 6 month old P48Cre;KrasG12D;Ikbke−/− mouse pancreas compared to age-matched P48Cre;Kras;G12D pancreas. (H) Subcellular localization of Gli1-GFP fusion protein in transfected 293T cells with (i–iii) and without (iv–vi) overlay with DAPI. While GLI1 is localized mainly in the cytoplasm without IKBKE expression (i, iv), IKBKE expression drives nuclear localization of GLI1 (ii, v). Expression of a kinase-dead version of IKBKE (K38A) causes cytoplasmic retention of GLI1 (iii, vi). (I) Co-expression of IKBKE, but not the kinases-dead version of IKBKE, with GLI1 synergistically increases GLI transcriptional activity as measured by Gli-BS Luciferase activity in 293T cells. IKBKE expression has no effect on the activity of Gli1-AHA, a mutant form of Gli1 that is constitutively localized to the nucleus. Error bars represent Standard Deviation. (J) Western blot analysis of nuclear and cytoplasmic fractions indicates that ectopic coexpression with wild type but not catalytically inactive (K38A) IKBKE leads to significant nuclear localization of GLI1. PCNA, and Tubulin are used as controls for nuclear (N) and cytoplasmic fractions (C) respectively. (K) Western blot analysis of nuclear and cytoplasmic fractions in Panc-1 cells indicates decrease in endogenous GLI1 nuclear localization after shRNA mediated knockdown of IKBKE. Statistical significance was determined using a Student’s two-tailed t-test. * P<0.05; ** P<0.01.