Figure 5.
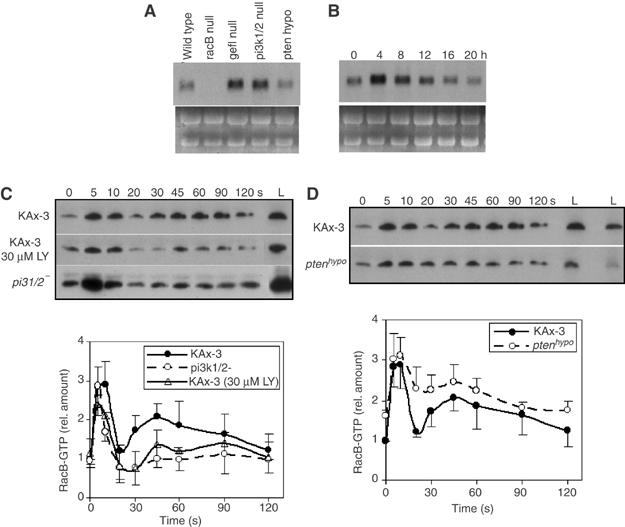
Regulation of RacB by the PI3K pathway. (A) RNA blot of total RNA isolated from vegetative wild-type, racB null, racgef1 null, pi3k1/2 null, and PTEN hypomorphic cells. (B) Developmental RNA blot of RacB RNA. Total cell RNA was isolated from developing wild-type cells plated on non-nutrient agar at the indicated times. 0, vegetatively growing cells; 4 h; early/mid aggregation; 8 h, mound stage; 12 h, tipped aggregates; 20 h, mature fruiting body. (Bush et al (1993) found RacB transcripts present at the same level in vegetative and 4 h starved cells.) (C, D) RacB-GTP in stimulated wild-type cells, wild-type cells pretreated with LY294002, and pi3k1/2 null cells (C) or wild-type and PTEN hypomorphic cells (D). Lanes labeled ‘L' show the levels of total myc-RacB in the same volume of each lysate. The amount of RacB-GTP was quantified by densitometry of developed Western blot films in four independent experiments (lower panel). The wild-type level at 0 s was set at 1.0. The levels of RacB-GTP in a specific mutant strain were normalized to RacB expression in wild-type cells by dividing the total amount of RacB in the mutant by that of wild-type cells. Error bars indicate standard deviation.
