Figure 7.
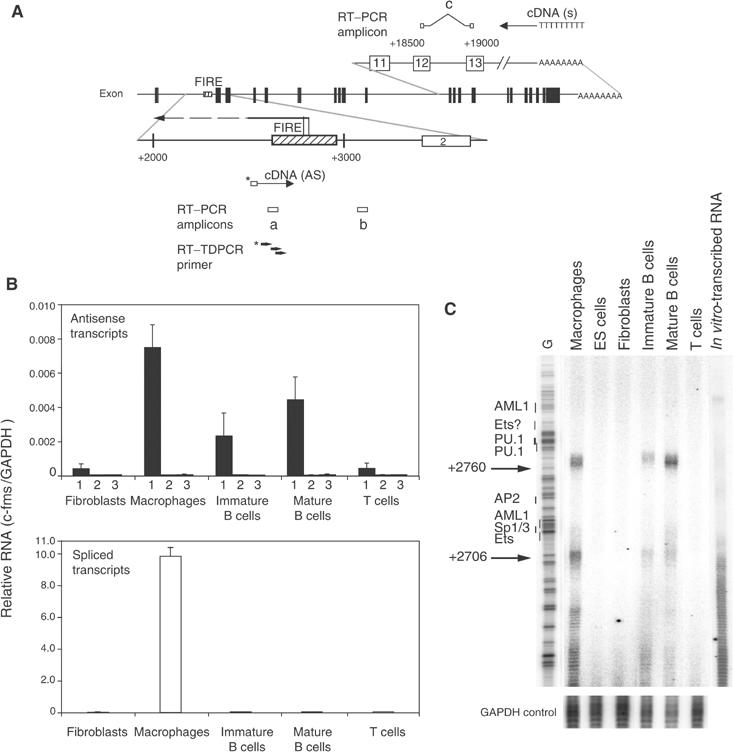
Antisense (AS) RNA starting at FIRE is expressed in macrophages and B cells. (A) Schematic representation of the position of primers used to carry out cDNA synthesis as well as real-time PCR amplicons. cDNA specific for AS RNA was synthesized from a biotinylated primer (indicated below the map), whereas an oligo (dT) primer was used to detect spliced transcripts (indicated above the map). Primer sets (a) (FIRE06) and (b) (3′-FIRE) were used to detect AS RNA transcribed from FIRE and from downstream of FIRE, respectively. Primer set (a) (c-fms QPCR), which is located between exons 12 and 13, was used to detect spliced sense transcripts. Black boxes in the map and numbered white boxes in the top and bottom map represent c-fms exons. Dashed boxes in the bottom row represent FIRE sequences. The numbers indicate the nucleotide position relative to the ATG start codon. The localization of the AS transcript is indicated as a horizontal arrow and the two AS transcription start sites mapped in (C) are indicated. (B) Expression of sense and AS RNA at the c-fms locus as assayed by real-time PCR. The top panel represents signals obtained with primer pair (a) measuring AS RNA originating from FIRE (1), signals from genomic DNA contamination (2) (assays with primer pair (a) but without reverse transcriptase) and signals obtained with primer pair (b) (3). The bottom panel represents spliced sense transcripts detected by primer pair (c). Bars represent mean value±s.d. of two to four independent experiments analysed in duplicate. (C) Determination of the start site of the AS RNA within FIRE. RT–TDPCR was performed as described in Materials and methods using RNA prepared from the indicated cell types and from in vitro-synthesized RNA as a control for cDNA synthesis and amplification artefacts. An RT–TDPCR reaction examining the GAPDH gene was performed as internal control. A sequence reaction was run on the gel alongside the RT–TDPCR samples and the positions of transcription factor-binding sites within FIRE are indicated. This result was confirmed in two independently performed experiments.
